MLL-Rearranged Acute Lymphoblastic Leukemias Activate BCL-2 through H3K79 Methylation and Are Sensitive to the BCL-2-Specific Antagonist ABT-199
- PMID: 26711339
- PMCID: PMC4700051
- DOI: 10.1016/j.celrep.2015.12.003
MLL-Rearranged Acute Lymphoblastic Leukemias Activate BCL-2 through H3K79 Methylation and Are Sensitive to the BCL-2-Specific Antagonist ABT-199
Abstract
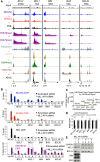
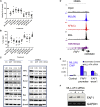
Targeted therapies designed to exploit specific molecular pathways in aggressive cancers are an exciting area of current research. Mixed Lineage Leukemia (MLL) mutations such as the t(4;11) translocation cause aggressive leukemias that are refractory to conventional treatment. The t(4;11) translocation produces an MLL/AF4 fusion protein that activates key target genes through both epigenetic and transcriptional elongation mechanisms. In this study, we show that t(4;11) patient cells express high levels of BCL-2 and are highly sensitive to treatment with the BCL-2-specific BH3 mimetic ABT-199. We demonstrate that MLL/AF4 specifically upregulates the BCL-2 gene but not other BCL-2 family members via DOT1L-mediated H3K79me2/3. We use this information to show that a t(4;11) cell line is sensitive to a combination of ABT-199 and DOT1L inhibitors. In addition, ABT-199 synergizes with standard induction-type therapy in a xenotransplant model, advocating for the introduction of ABT-199 into therapeutic regimens for MLL-rearranged leukemias.
Keywords: DOT1L; H3K79 methylation; MLL/AF4; apoptosis pathways; bcl-2 family members; leukemias.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Andersson A.K., Ma J., Wang J., Chen X., Gedman A.L., Dang J., Nakitandwe J., Holmfeldt L., Parker M., Easton J., St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat. Genet. 2015;47:330–337. - PMC - PubMed
-
- Beldjord K., Chevret S., Asnafi V., Huguet F., Boulland M.L., Leguay T., Thomas X., Cayuela J.M., Grardel N., Chalandon Y., Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) Oncogenetics and minimal residual disease are independent outcome predictors in adult patients with acute lymphoblastic leukemia. Blood. 2014;123:3739–3749. - PubMed
-
- Bursen A., Schwabe K., Rüster B., Henschler R., Ruthardt M., Dingermann T., Marschalek R. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood. 2010;115:3570–3579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

