Compartmentalization and Functionality of Nuclear Disorder: Intrinsic Disorder and Protein-Protein Interactions in Intra-Nuclear Compartments
- PMID: 26712748
- PMCID: PMC4730271
- DOI: 10.3390/ijms17010024
Compartmentalization and Functionality of Nuclear Disorder: Intrinsic Disorder and Protein-Protein Interactions in Intra-Nuclear Compartments
Abstract
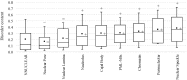
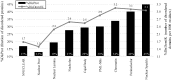
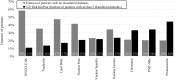
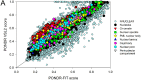
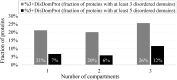
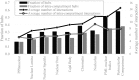
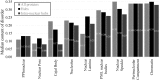
The cell nucleus contains a number of membrane-less organelles or intra-nuclear compartments. These compartments are dynamic structures representing liquid-droplet phases which are only slightly denser than the bulk intra-nuclear fluid. They possess different functions, have diverse morphologies, and are typically composed of RNA (or, in some cases, DNA) and proteins. We analyzed 3005 mouse proteins localized in specific intra-nuclear organelles, such as nucleolus, chromatin, Cajal bodies, nuclear speckles, promyelocytic leukemia (PML) nuclear bodies, nuclear lamina, nuclear pores, and perinuclear compartment and compared them with ~29,863 non-nuclear proteins from mouse proteome. Our analysis revealed that intrinsic disorder is enriched in the majority of intra-nuclear compartments, except for the nuclear pore and lamina. These compartments are depleted in proteins that lack disordered domains and enriched in proteins that have multiple disordered domains. Moonlighting proteins found in multiple intra-nuclear compartments are more likely to have multiple disordered domains. Protein-protein interaction networks in the intra-nuclear compartments are denser and include more hubs compared to the non-nuclear proteins. Hubs in the intra-nuclear compartments (except for the nuclear pore) are enriched in disorder compared with non-nuclear hubs and non-nuclear proteins. Therefore, our work provides support to the idea of the functional importance of intrinsic disorder in the cell nucleus and shows that many proteins associated with sub-nuclear organelles in nuclei of mouse cells are enriched in disorder. This high level of disorder in the mouse nuclear proteins defines their ability to serve as very promiscuous binders, possessing both large quantities of potential disorder-based interaction sites and the ability of a single such site to be involved in a large number of interactions.
Keywords: DNA-binding protein; RNA-binding protein; cell nucleus; intra-nuclear compartments; intrinsically disordered protein; membrane-less organelles.
Figures



References
-
- Tripathi V., Prasanth K.V. eLS. John Wiley & Sons Ltd.; Chichester, UK: 2011. Cell nucleus.
-
- Hancock R., Jeon K. New Models of the Cell Nucleus: Crowding, Entropic Forces, Phase Separation, and Fractals. Academic Press; Amsterdam, The Netherlands; Boston, MA, USA; Heidelberg, Germany; London, UK; New York, NY, USA; Oxford, UK; Paris, France; San Diego, CA, USA; San Francisco, CA, USA; Singapoe; Sydney, Austrilia; Tokyo, Jaoan: 2014. p. 512.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

