ISC, a Novel Group of Bacterial and Archaeal DNA Transposons That Encode Cas9 Homologs
- PMID: 26712934
- PMCID: PMC4810608
- DOI: 10.1128/JB.00783-15
ISC, a Novel Group of Bacterial and Archaeal DNA Transposons That Encode Cas9 Homologs
Abstract
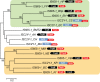
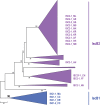
Bacterial genomes encode numerous homologs of Cas9, the effector protein of the type II CRISPR-Cas systems. The homology region includes the arginine-rich helix and the HNH nuclease domain that is inserted into the RuvC-like nuclease domain. These genes, however, are not linked to cas genes or CRISPR. Here, we show that Cas9 homologs represent a distinct group of nonautonomous transposons, which we denote ISC (insertion sequences Cas9-like). We identify many diverse families of full-length ISC transposons and demonstrate that their terminal sequences (particularly 3' termini) are similar to those of IS605 superfamily transposons that are mobilized by the Y1 tyrosine transposase encoded by the TnpA gene and often also encode the TnpB protein containing the RuvC-like endonuclease domain. The terminal regions of the ISC and IS605 transposons contain palindromic structures that are likely recognized by the Y1 transposase. The transposons from these two groups are inserted either exactly in the middle or upstream of specific 4-bp target sites, without target site duplication. We also identify autonomous ISC transposons that encode TnpA-like Y1 transposases. Thus, the nonautonomous ISC transposons could be mobilized in trans either by Y1 transposases of other, autonomous ISC transposons or by Y1 transposases of the more abundant IS605 transposons. These findings imply an evolutionary scenario in which the ISC transposons evolved from IS605 family transposons, possibly via insertion of a mobile group II intron encoding the HNH domain, and Cas9 subsequently evolved via immobilization of an ISC transposon.
Importance: Cas9 endonucleases, the effectors of type II CRISPR-Cas systems, represent the new generation of genome-engineering tools. Here, we describe in detail a novel family of transposable elements that encode the likely ancestors of Cas9 and outline the evolutionary scenario connecting different varieties of these transposons and Cas9.
Copyright © 2016 Kapitonov et al.
Figures




References
-
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7. doi: 10.1186/1745-6150-1-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

