Brain Transcriptomic Response to Social Eavesdropping in Zebrafish (Danio rerio)
- PMID: 26713440
- PMCID: PMC4700982
- DOI: 10.1371/journal.pone.0145801
Brain Transcriptomic Response to Social Eavesdropping in Zebrafish (Danio rerio)
Abstract
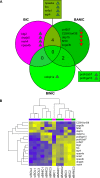
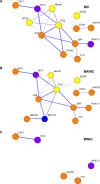
Public information is widely available at low cost to animals living in social groups. For instance, bystanders may eavesdrop on signaling interactions between conspecifics and use it to adapt their subsequent behavior towards the observed individuals. This social eavesdropping ability is expected to require specialized mechanisms such as social attention, which selects social information available for learning. To begin exploring the genetic basis of social eavesdropping, we used a previously established attention paradigm in the lab to study the brain gene expression profile of male zebrafish (Danio rerio) in relation to the attention they paid towards conspecifics involved or not involved in agonistic interactions. Microarray gene chips were used to characterize their brain transcriptomes based on differential expression of single genes and gene sets. These analyses were complemented by promoter region-based techniques. Using data from both approaches, we further drafted protein interaction networks. Our results suggest that attentiveness towards conspecifics, whether interacting or not, activates pathways linked to neuronal plasticity and memory formation. The network analyses suggested that fos and jun are key players on this response, and that npas4a, nr4a1 and egr4 may also play an important role. Furthermore, specifically observing fighting interactions further triggered pathways associated to a change in the alertness status (dnajb5) and to other genes related to memory formation (btg2, npas4b), which suggests that the acquisition of eavesdropped information about social relationships activates specific processes on top of those already activated just by observing conspecifics.
Conflict of interest statement
Figures


References
-
- McGregor PK. Signalling in Territorial Systems: A Context for Individual Identification, Ranging and Eavesdropping. Philos Trans R Soc B Biol Sci. 1993; 340: 237–244. 10.1098/rstb.1993.0063 - DOI
-
- Peake TM. Eavesdropping in communication networks In: McGregor P, editor. Anim Commun networks. Cambridge: Cambridge University Press; 2005; pp. 13–37.
-
- Byrne RW, Whiten A. Machiavellian Intelligence: Social Expertise and the Evolution of Intellect in Monkeys, Apes, and Humans Oxford: Oxford University Press; 1989. p. 599 10.1016/0003-3472(89)90054-7 - DOI
-
- Humphrey NK. The social function of intellect In: Bateson P, Hinde R, editors. Growing points in ethology. Cambridge: Cambridge University Press; 1976. pp. 303–317.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

