Modeling-Enabled Characterization of Novel NLRX1 Ligands
- PMID: 26714018
- PMCID: PMC4694766
- DOI: 10.1371/journal.pone.0145420
Modeling-Enabled Characterization of Novel NLRX1 Ligands
Abstract
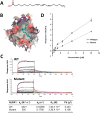
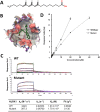
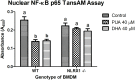
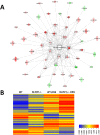
Nucleotide-binding domain and leucine-rich repeat containing (NLR) family are intracellular sentinels of cytosolic homeostasis that orchestrate immune and inflammatory responses in infectious and immune-mediated diseases. NLRX1 is a mitochondrial-associated NOD-like receptor involved in the modulation of immune and metabolic responses. This study utilizes molecular docking approaches to investigate the structure of NLRX1 and experimentally assesses binding to naturally occurring compounds from several natural product and lipid databases. Screening of compound libraries predicts targeting of NLRX1 by conjugated trienes, polyketides, prenol lipids, sterol lipids, and coenzyme A-containing fatty acids for activating the NLRX1 pathway. The ligands of NLRX1 were identified by docking punicic acid (PUA), eleostearic acid (ESA), and docosahexaenoic acid (DHA) to the C-terminal fragment of the human NLRX1 (cNLRX1). Their binding and that of positive control RNA to cNLRX1 were experimentally determined by surface plasmon resonance (SPR) spectroscopy. In addition, the ligand binding sites of cNLRX1 were predicted in silico and validated experimentally. Target mutagenesis studies demonstrate that mutation of 4 critical residues ASP677, PHE680, PHE681, and GLU684 to alanine resulted in diminished affinity of PUA, ESA, and DHA to NLRX1. Consistent with the regulatory actions of NLRX1 on the NF-κB pathway, treatment of bone marrow derived macrophages (BMDM)s with PUA and DHA suppressed NF-κB activity in a NLRX1 dependent mechanism. In addition, a series of pre-clinical efficacy studies were performed using a mouse model of dextran sodium sulfate (DSS)-induced colitis. Our findings showed that the regulatory function of PUA on colitis is NLRX1 dependent. Thus, we identified novel small molecules that bind to NLRX1 and exert anti-inflammatory actions.
Conflict of interest statement
Figures


References
-
- Franchi L, McDonald C, Kanneganti T-D, Amer A, Núñez G. Nucleotide-binding oligomerization domain-like receptors: intracellular pattern recognition molecules for pathogen detection and host defense. The Journal of Immunology. 2006;177(6):3507–13. - PubMed
-
- Harton JA, Linhoff MW, Zhang J, Ting JP-Y. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. The Journal of Immunology. 2002;169(8):4088–93. - PubMed
-
- Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20(3):319–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

