Protein translocation channel of mitochondrial inner membrane and matrix-exposed import motor communicate via two-domain coupling protein
- PMID: 26714107
- PMCID: PMC4749553
- DOI: 10.7554/eLife.11897
Protein translocation channel of mitochondrial inner membrane and matrix-exposed import motor communicate via two-domain coupling protein
Abstract
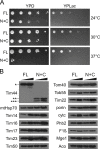
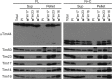
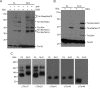
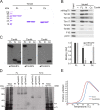
The majority of mitochondrial proteins are targeted to mitochondria by N-terminal presequences and use the TIM23 complex for their translocation across the mitochondrial inner membrane. During import, translocation through the channel in the inner membrane is coupled to the ATP-dependent action of an Hsp70-based import motor at the matrix face. How these two processes are coordinated remained unclear. We show here that the two domain structure of Tim44 plays a central role in this process. The N-terminal domain of Tim44 interacts with the components of the import motor, whereas its C-terminal domain interacts with the translocation channel and is in contact with translocating proteins. Our data suggest that the translocation channel and the import motor of the TIM23 complex communicate through rearrangements of the two domains of Tim44 that are stimulated by translocating proteins.
Keywords: E. coli; Hsp70; S. cerevisiae; TIM23; biochemistry; cell biology; mitochondria; protein translocation/sorting.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



Similar articles
-
Architecture of the TIM23 inner mitochondrial translocon and interactions with the matrix import motor.J Biol Chem. 2014 Oct 10;289(41):28689-96. doi: 10.1074/jbc.M114.588152. Epub 2014 Aug 25. J Biol Chem. 2014. PMID: 25157107 Free PMC article.
-
Dual interaction of scaffold protein Tim44 of mitochondrial import motor with channel-forming translocase subunit Tim23.Elife. 2017 Apr 25;6:e23609. doi: 10.7554/eLife.23609. Elife. 2017. PMID: 28440746 Free PMC article.
-
Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase.Mol Biol Cell. 2008 Jun;19(6):2642-9. doi: 10.1091/mbc.e07-12-1226. Epub 2008 Apr 9. Mol Biol Cell. 2008. PMID: 18400944 Free PMC article.
-
On the mechanism of preprotein import by the mitochondrial presequence translocase.Biochim Biophys Acta. 2010 Jun;1803(6):732-9. doi: 10.1016/j.bbamcr.2010.01.013. Epub 2010 Jan 25. Biochim Biophys Acta. 2010. PMID: 20100523 Review.
-
How to get to the other side of the mitochondrial inner membrane - the protein import motor.Biol Chem. 2020 May 26;401(6-7):723-736. doi: 10.1515/hsz-2020-0106. Biol Chem. 2020. PMID: 32142474 Review.
Cited by
-
Two mitochondrial HMG-box proteins, Cim1 and Abf2, antagonistically regulate mtDNA copy number in Saccharomyces cerevisiae.Nucleic Acids Res. 2023 Nov 27;51(21):11813-11835. doi: 10.1093/nar/gkad849. Nucleic Acids Res. 2023. PMID: 37850632 Free PMC article.
-
Homologue replacement in the import motor of the mitochondrial inner membrane of trypanosomes.Elife. 2020 Feb 27;9:e52560. doi: 10.7554/eLife.52560. Elife. 2020. PMID: 32105215 Free PMC article.
-
Molecular basis of resistance to the microtubule-depolymerizing antitumor compound plocabulin.Sci Rep. 2018 Jun 5;8(1):8616. doi: 10.1038/s41598-018-26736-3. Sci Rep. 2018. PMID: 29872155 Free PMC article.
-
SAMM50 acts with p62 in piecemeal basal- and OXPHOS-induced mitophagy of SAM and MICOS components.J Cell Biol. 2021 Aug 2;220(8):e202009092. doi: 10.1083/jcb.202009092. Epub 2021 May 26. J Cell Biol. 2021. PMID: 34037656 Free PMC article.
-
Targeting and Insertion of Membrane Proteins in Mitochondria.Front Cell Dev Biol. 2021 Dec 24;9:803205. doi: 10.3389/fcell.2021.803205. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 35004695 Free PMC article. Review.
References
-
- Bonora E, Evangelisti C, Bonichon F, Tallini G, Romeo G. Novel germline variants identified in the inner mitochondrial membrane transporter TIMM44 and their role in predisposition to oncocytic thyroid carcinomas. British Journal of Cancer. 2006;95:1529–1536. doi: 10.1038/sj.bjc.6603455. - DOI - PMC - PubMed
-
- Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

