A general strategy to construct small molecule biosensors in eukaryotes
- PMID: 26714111
- PMCID: PMC4739774
- DOI: 10.7554/eLife.10606
A general strategy to construct small molecule biosensors in eukaryotes
Abstract
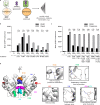
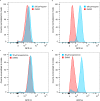
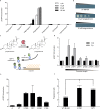
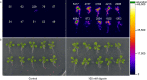
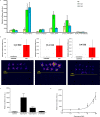
Biosensors for small molecules can be used in applications that range from metabolic engineering to orthogonal control of transcription. Here, we produce biosensors based on a ligand-binding domain (LBD) by using a method that, in principle, can be applied to any target molecule. The LBD is fused to either a fluorescent protein or a transcriptional activator and is destabilized by mutation such that the fusion accumulates only in cells containing the target ligand. We illustrate the power of this method by developing biosensors for digoxin and progesterone. Addition of ligand to yeast, mammalian, or plant cells expressing a biosensor activates transcription with a dynamic range of up to ~100-fold. We use the biosensors to improve the biotransformation of pregnenolone to progesterone in yeast and to regulate CRISPR activity in mammalian cells. This work provides a general methodology to develop biosensors for a broad range of molecules in eukaryotes.
Keywords: a. thaliana; s. cerevisiae; CRISPR; biochemistry; biophysics; biosensors; human; metabolic engineering; structural biology.
Conflict of interest statement
JF, BWJ, CET, DJM, RC, XR, GMC, SF, DB: Harvard University has filed a provisional patent on this work.
The other authors declare that no competing interests exist.
Figures







References
-
- Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret J-C, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

