Complex Genotype Mixtures Analyzed by Deep Sequencing in Two Different Regions of Hepatitis B Virus
- PMID: 26714168
- PMCID: PMC4695080
- DOI: 10.1371/journal.pone.0144816
Complex Genotype Mixtures Analyzed by Deep Sequencing in Two Different Regions of Hepatitis B Virus
Abstract
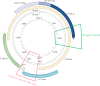
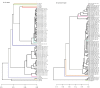
This study assesses the presence and outcome of genotype mixtures in the polymerase/surface and X/preCore regions of the HBV genome in patients with chronic hepatitis B virus (HBV) infection. Thirty samples from ten chronic hepatitis B patients were included. The polymerase/surface and X/preCore regions were analyzed by deep sequencing (UDPS) in the first available sample at diagnosis, a pre-treatment sample, and a sample while under treatment. HBV genotype was determined by phylogenesis. Quasispecies complexity was evaluated by mutation frequency and nucleotide diversity. The polymerase/surface and X/preCore regions were validated for genotyping from 113 GenBank reference sequences. UDPS yielded a median of 10,960 sequences per sample (IQR 16,645) in the polymerase/surface region and 11,595 sequences per sample (IQR 14,682) in X/preCore. Genotype mixtures were more common in X/preCore (90%) than in polymerase/surface (30%) (p<0.001). On X/preCore genotyping, all samples were genotype A, whereas polymerase/surface yielded genotypes A (80%), D (16.7%), and F (3.3%) (p = 0.036). Genotype changes in polymerase/surface were observed in four patients during natural quasispecies dynamics and in two patients during treatment. There were no genotype changes in X/preCore. Quasispecies complexity was higher in X/preCore than in polymerase/surface (p = 0.004). The results provide evidence of genotype mixtures and differential genotype proportions in the polymerase/surface and X/preCore regions. The genotype dynamics in HBV infection and the different patterns of quasispecies complexity in the HBV genome suggest a new paradigm for HBV genotype classification.
Conflict of interest statement
Figures

Similar articles
-
Comparative study of genotype B and C hepatitis B virus-induced chronic hepatitis in relation to the basic core promoter and precore mutations.J Gastroenterol Hepatol. 2005 Mar;20(3):441-9. doi: 10.1111/j.1440-1746.2004.03572.x. J Gastroenterol Hepatol. 2005. PMID: 15740490
-
Analysis of ultra-deep pyrosequencing and cloning based sequencing of the basic core promoter/precore/core region of hepatitis B virus using newly developed bioinformatics tools.PLoS One. 2014 Apr 16;9(4):e95377. doi: 10.1371/journal.pone.0095377. eCollection 2014. PLoS One. 2014. PMID: 24740330 Free PMC article.
-
Nucleic acid sequence analysis of basal core promoter/precore/core region of hepatitis B virus isolated from chronic carriers of the virus from Kolkata, eastern India: low frequency of mutation in the precore region.Intervirology. 2005;48(6):389-99. doi: 10.1159/000086066. Intervirology. 2005. PMID: 16024943
-
Quasispecies structure, cornerstone of hepatitis B virus infection: mass sequencing approach.World J Gastroenterol. 2013 Nov 7;19(41):6995-7023. doi: 10.3748/wjg.v19.i41.6995. World J Gastroenterol. 2013. PMID: 24222943 Free PMC article. Review.
-
Next-generation sequencing for the diagnosis of hepatitis B: current status and future prospects.Expert Rev Mol Diagn. 2021 Apr;21(4):381-396. doi: 10.1080/14737159.2021.1913055. Epub 2021 Apr 21. Expert Rev Mol Diagn. 2021. PMID: 33880971 Review.
Cited by
-
Analysis of hepatitis B virus-mixed genotype infection by ultra deep pyrosequencing in Sudanese patients, 2015-2016.Infection. 2019 Oct;47(5):793-803. doi: 10.1007/s15010-019-01306-5. Epub 2019 Apr 8. Infection. 2019. PMID: 30963405
-
A New Method for Next-Generation Sequencing of the Full Hepatitis B Virus Genome from A Clinical Specimen: Impact for Virus Genotyping.Microorganisms. 2020 Sep 11;8(9):1391. doi: 10.3390/microorganisms8091391. Microorganisms. 2020. PMID: 32932752 Free PMC article.
-
Hepatitis B Virus Variants with Multiple Insertions and/or Deletions in the X Open Reading Frame 3' End: Common Members of Viral Quasispecies in Chronic Hepatitis B Patients.Biomedicines. 2022 May 21;10(5):1194. doi: 10.3390/biomedicines10051194. Biomedicines. 2022. PMID: 35625929 Free PMC article.
-
Insights From Deep Sequencing of the HBV Genome-Unique, Tiny, and Misunderstood.Gastroenterology. 2019 Jan;156(2):384-399. doi: 10.1053/j.gastro.2018.07.058. Epub 2018 Sep 27. Gastroenterology. 2019. PMID: 30268787 Free PMC article. Review.
-
An Oxford Nanopore Technology-Based Hepatitis B Virus Sequencing Protocol Suitable For Genomic Surveillance Within Clinical Diagnostic Settings.medRxiv [Preprint]. 2024 Jan 25:2024.01.19.24301519. doi: 10.1101/2024.01.19.24301519. medRxiv. 2024. Update in: Int J Mol Sci. 2024 Oct 31;25(21):11702. doi: 10.3390/ijms252111702. PMID: 38293032 Free PMC article. Updated. Preprint.
References
-
- WHO. WHO | Hepatitis B [Internet]. World Health Organization; 2015 [Accessed 4 Jun 2015]. Available: http://www.who.int/mediacentre/factsheets/fs204/en/
-
- Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, et al. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69 (Pt 10: 2575–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

