The biogeography of polymicrobial infection
- PMID: 26714431
- PMCID: PMC5116812
- DOI: 10.1038/nrmicro.2015.8
The biogeography of polymicrobial infection
Abstract
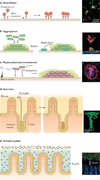
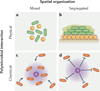
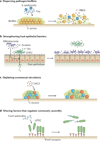
Microbial communities are spatially organized in both the environment and the human body. Although patterns exhibited by these communities are described by microbial biogeography, this discipline has previously only considered large-scale, global patterns. By contrast, the fine-scale positioning of a pathogen within an infection site can greatly alter its virulence potential. In this Review, we highlight the importance of considering spatial positioning in the study of polymicrobial infections and discuss targeting biogeography as a therapeutic strategy.
Figures
References
-
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. - PubMed
-
- Kaufmann SH, Schaible UE. 100th anniversary of Robert Koch’s Nobel Prize for the discovery of the tubercle bacillus. Trends Microbiol. 2005;13:469–475. - PubMed
-
- Smith H. The role of microbial interactions in infectious disease. Phil. Trans. R. Soc. Lond. B. Biol. Sci. 1982;297:551–561. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

