Effect of roughage on rumen microbiota composition in the efficient feed converter and sturdy Indian Jaffrabadi buffalo (Bubalus bubalis)
- PMID: 26714477
- PMCID: PMC4696265
- DOI: 10.1186/s12864-015-2340-4
Effect of roughage on rumen microbiota composition in the efficient feed converter and sturdy Indian Jaffrabadi buffalo (Bubalus bubalis)
Abstract
Background: The rumen microbiota functions as an effective system for conversion of dietary feed to microbial proteins and volatile fatty acids. In the present study, metagenomic approach was applied to elucidate the buffalo rumen microbiome of Jaffrabadi buffalo adapted to varied dietary treatments with the hypothesis that the microbial diversity and subsequent in the functional capacity will alter with diet change and enhance our knowledge of effect of microbe on host physiology. Eight adult animals were gradually adapted to an increasing roughage diet (4 animals each with green and dry roughage) containing 50:50 (J1), 75:25 (J2) and 100:0 (J3) roughage to concentrate proportion for 6 weeks. Metagenomic sequences of solid (fiber adherent microbiota) and liquid (fiber free microbiota) fractions obtained using Ion Torrent PGM platform were analyzed using MG-RAST server and CAZymes approach.
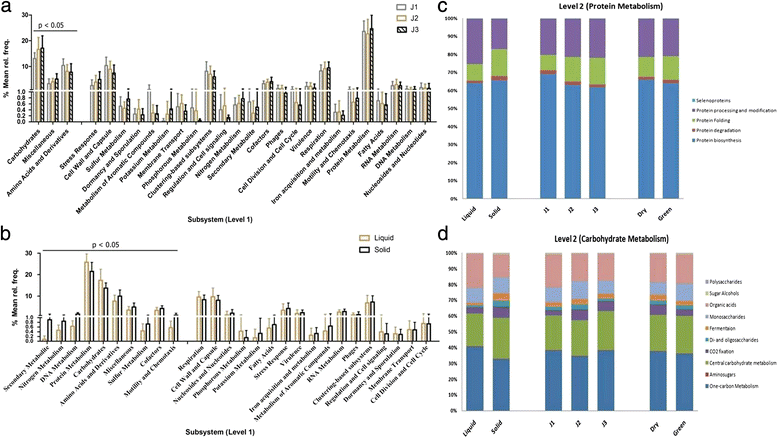
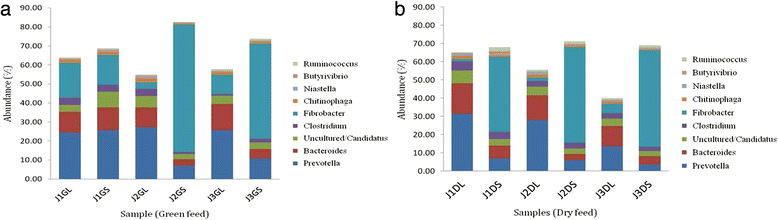
Results: Taxonomic analysis revealed that Bacteroidetes was the most abundant phylum followed by Firmicutes, Fibrobacter and Proteobacteria. Functional analysis revealed protein (25-30 %) and carbohydrate (15-20 %) metabolism as the dominant categories. Principal component analysis demonstrated that roughage proportion, fraction of rumen and type of forage affected rumen microbiome at taxonomic as well as functional level. Rumen metabolite study revealed that rumen fluid nitrogen content reduced in high roughage diet fed animals and pathway analysis showed reduction in the genes coding enzymes involved in methanogenesis pathway. CAZyme annotation revealed the abundance of genes encoding glycoside hydrolases (GH), with the GH3 family most abundant followed by GH2 and GH13 in all samples.
Conclusions: Results reveals that high roughage diet feed improved microbial protein synthesis and reduces methane emission. CAZyme analysis indicated the importance of microbiome in feed component digestion for fulfilling energy requirements of the host. The findings help determine the role of rumen microbes in plant polysaccharide breakdown and in developing strategies to maximize productivity in ruminants.
Figures




Similar articles
-
Understanding the alteration in rumen microbiome and CAZymes profile with diet and host through comparative metagenomic approach.Arch Microbiol. 2019 Dec;201(10):1385-1397. doi: 10.1007/s00203-019-01706-z. Epub 2019 Jul 23. Arch Microbiol. 2019. PMID: 31338542
-
Functional gene profiling through metaRNAseq approach reveals diet-dependent variation in rumen microbiota of buffalo (Bubalus bubalis).Anaerobe. 2017 Apr;44:106-116. doi: 10.1016/j.anaerobe.2017.02.021. Epub 2017 Feb 27. Anaerobe. 2017. PMID: 28246035
-
Microbial and Carbohydrate Active Enzyme profile of buffalo rumen metagenome and their alteration in response to variation in the diet.Gene. 2014 Jul 15;545(1):88-94. doi: 10.1016/j.gene.2014.05.003. Epub 2014 May 2. Gene. 2014. PMID: 24797613
-
The early impact of genomics and metagenomics on ruminal microbiology.Annu Rev Anim Biosci. 2015;3:447-65. doi: 10.1146/annurev-animal-022114-110705. Epub 2014 Oct 9. Annu Rev Anim Biosci. 2015. PMID: 25387109 Review.
-
Symposium review: Mining metagenomic and metatranscriptomic data for clues about microbial metabolic functions in ruminants.J Dairy Sci. 2018 Jun;101(6):5605-5618. doi: 10.3168/jds.2017-13356. Epub 2017 Dec 21. J Dairy Sci. 2018. PMID: 29274958 Review.
Cited by
-
Impact of virginiamycin on the ruminal microbiota of feedlot cattle.Can J Vet Res. 2024 Oct;88(4):114-122. Can J Vet Res. 2024. PMID: 39355681 Free PMC article.
-
Changes in the bacterial community colonizing extracted and non-extracted tannin-rich plants in the rumen of dromedary camels.PLoS One. 2023 Mar 10;18(3):e0282889. doi: 10.1371/journal.pone.0282889. eCollection 2023. PLoS One. 2023. PMID: 36897876 Free PMC article.
-
Lignocelluloytic activities and composition of bacterial community in the camel rumen.AIMS Microbiol. 2021 Sep 24;7(3):354-367. doi: 10.3934/microbiol.2021022. eCollection 2021. AIMS Microbiol. 2021. PMID: 34708177 Free PMC article.
-
Comparative Rumen Metagenome and CAZyme Profiles in Cattle and Buffaloes: Implications for Methane Yield and Rumen Fermentation on a Common Diet.Microorganisms. 2023 Dec 27;12(1):47. doi: 10.3390/microorganisms12010047. Microorganisms. 2023. PMID: 38257874 Free PMC article.
-
Stability Assessment of the Rumen Bacterial and Archaeal Communities in Dairy Cows Within a Single Lactation and Its Association With Host Phenotype.Front Microbiol. 2021 Apr 6;12:636223. doi: 10.3389/fmicb.2021.636223. eCollection 2021. Front Microbiol. 2021. PMID: 33927700 Free PMC article.
References
-
- Goodacre R. Metabolomics of a superorganism. J Nutr. 2007;137(1 Suppl):259S–266S. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

