Characterization of Two Distinct Nucleosome Remodeling and Deacetylase (NuRD) Complex Assemblies in Embryonic Stem Cells
- PMID: 26714524
- PMCID: PMC4813707
- DOI: 10.1074/mcp.M115.053207
Characterization of Two Distinct Nucleosome Remodeling and Deacetylase (NuRD) Complex Assemblies in Embryonic Stem Cells
Abstract
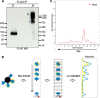
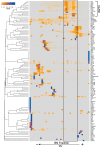
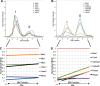
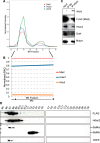
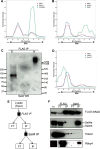
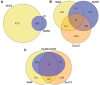
Pluripotency and self-renewal, the defining properties of embryonic stem cells, are brought about by transcriptional programs involving an intricate network of transcription factors and chromatin remodeling complexes. The Nucleosome Remodeling and Deacetylase (NuRD) complex plays a crucial and dynamic role in the regulation of stemness and differentiation. Several NuRD-associated factors have been reported but how they are organized has not been investigated in detail. Here, we have combined affinity purification and blue native polyacrylamide gel electrophoresis followed by protein identification by mass spectrometry and protein correlation profiling to characterize the topology of the NuRD complex. Our data show that in mouse embryonic stem cells the NuRD complex is present as two distinct assemblies of differing topology with different binding partners. Cell cycle regulator Cdk2ap1 and transcription factor Sall4 associate only with the higher mass NuRD assembly. We further establish that only isoform Sall4a, and not Sall4b, associates with NuRD. By contrast, Suz12, a component of the PRC2 Polycomb repressor complex, associates with the lower mass entity. In addition, we identify and validate a novel NuRD-associated protein, Wdr5, a regulatory subunit of the MLL histone methyltransferase complex, which associates with both NuRD entities. Bioinformatic analyses of published target gene sets of these chromatin binding proteins are in agreement with these structural observations. In summary, this study provides an interesting insight into mechanistic aspects of NuRD function in stem cell biology. The relevance of our work has broader implications because of the ubiquitous nature of the NuRD complex. The strategy described here can be more broadly applicable to investigate the topology of the multiple complexes an individual protein can participate in.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- Luo M., Ling T., Xie W., Sun H., Zhou Y., Zhu Q., Shen M., Zong L., Lyu G., Zhao Y., Ye T., Gu J., Tao W., Lu Z., and Grummt I. (2013) NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells 31, 1278–1286 - PubMed
-
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., and Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

