Adopting the rapamycin trapping assay to track the trafficking of murine MHC class I alleles, H-2K(b)
- PMID: 26714929
- PMCID: PMC4696223
- DOI: 10.1186/s12860-015-0077-1
Adopting the rapamycin trapping assay to track the trafficking of murine MHC class I alleles, H-2K(b)
Abstract
Background: In mammalian cells, the quality control (QC) of properly folded proteins is monitored in the early secretory pathway, particularly in the endoplasmic reticulum (ER). Several proteins, including our protein of interest, major histocompatibility complex class I (MHC class I), can bypass the first line of ER-QC and reside in post-ER compartments in an unfolded form. Such forms entail both monomeric and dimeric structures that are devoid of peptides and thus cannot fulfill the immunological function of antigen presentation at the cell surface. MHC class I structures become mature and properly folded once loaded with the appropriate peptides in the framework of the peptide loading complex (PLC). Despite the flood of information on the diverse trafficking behavior of different MHC class I alleles, there is still controversy on the actual trajectory followed by improperly folded murine MHC class I alleles, namely H-2Kb. In this study, we employ an in vitro rapamycin trapping assay, live cell imaging, and a biochemical COPII budding approach to further investigate the trafficking of H-2Kb beyond the level of the ER.
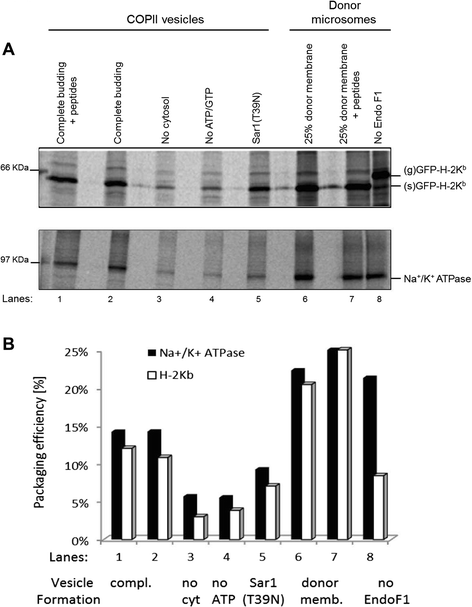
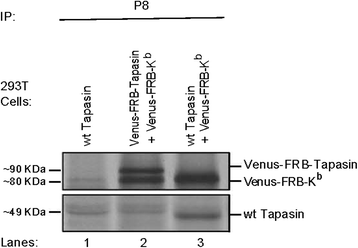
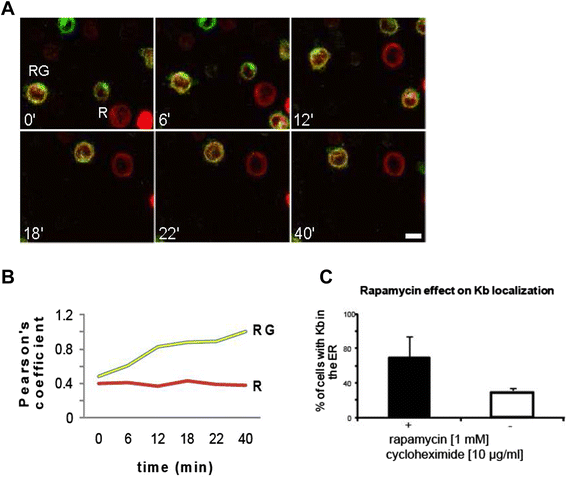
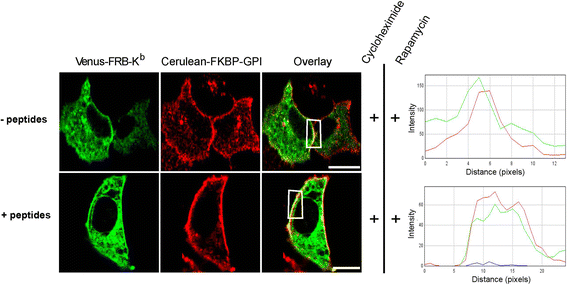
Results: We confirm the egress of H-2Kb in an unfolded form to a post-ER compartment from where they can cycle back to the ER. Deciphering the exact identity of the post-ER compartment by laser scanning microscopy did not only point to the existence of the ERGIC and cis-Golgi compartments as residency areas for unfolded proteins, but also to the involvement of an addional compartment, that lies in close proximity and possesses high resemblance to the aforementioned compartments. Interestingly, we were capable of showing using the same rapamycin trapping assay that H-2Kb can undergo a potential maturation event during their cycling; this is attained upon addition of peptides and trapping of accumulated post-ER molecules at the cell surface.
Conclusions: Our findings deepen the understanding of H-2Kb trafficking outside the ER and pave the way to decipher the role and the trafficking of certain PLC chaperones, such as tapasin, throughout H-2K(b) post-ER QC. Finally, we demonstrate the plausible usage of the rapamycin assay to assess the trafficking of defected proteins especially in diseases and under therapeutic studies.
Figures




Similar articles
-
Release from endoplasmic reticulum matrix proteins controls cell surface transport of MHC class I molecules.Traffic. 2015 Jun;16(6):591-603. doi: 10.1111/tra.12279. Epub 2015 Apr 16. Traffic. 2015. PMID: 25753898
-
The transporter associated with antigen processing (TAP) is active in a post-ER compartment.J Cell Sci. 2010 Dec 15;123(Pt 24):4271-9. doi: 10.1242/jcs.060632. Epub 2010 Nov 23. J Cell Sci. 2010. PMID: 21098634
-
Probing for membrane domains in the endoplasmic reticulum: retention and degradation of unassembled MHC class I molecules.Mol Biol Cell. 2002 May;13(5):1566-81. doi: 10.1091/mbc.01-07-0322. Mol Biol Cell. 2002. PMID: 12006653 Free PMC article.
-
How to Avoid a No-Deal ER Exit.Cells. 2019 Sep 7;8(9):1051. doi: 10.3390/cells8091051. Cells. 2019. PMID: 31500301 Free PMC article. Review.
-
Control of MHC class I traffic from the endoplasmic reticulum by cellular chaperones and viral anti-chaperones.Traffic. 2000 Apr;1(4):306-11. doi: 10.1034/j.1600-0854.2000.010403.x. Traffic. 2000. PMID: 11208115 Review.
Cited by
-
The temporal expression pattern of classical MHC class I in sleep-restricted mice: Generalizations and broader implications.Brain Behav Immun Health. 2024 Mar 11;37:100751. doi: 10.1016/j.bbih.2024.100751. eCollection 2024 May. Brain Behav Immun Health. 2024. PMID: 38511151 Free PMC article.
References
-
- Raine T, Brown D, Bowness P, Hill Gaston JS, Moffett A, Trowsdale J, et al. Consistent patterns of expression of HLA class I free heavy chains in healthy individuals and raised expression in spondyloarthropathy patients point to physiological and pathological roles. Rheumatol. 2006;45(11):1338–44. doi: 10.1093/rheumatology/kel305. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

