Isolation of a Hypomorphic skn-1 Allele That Does Not Require a Balancer for Maintenance
- PMID: 26715089
- PMCID: PMC4777118
- DOI: 10.1534/g3.115.023010
Isolation of a Hypomorphic skn-1 Allele That Does Not Require a Balancer for Maintenance
Abstract
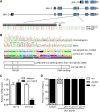
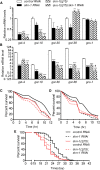
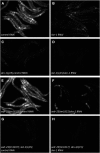
In Caenorhabditis elegans, the transcription factor SKN-1 has emerged as a central coordinator of stress responses and longevity, increasing the need for genetic tools to study its regulation and function. However, current loss-of-function alleles cause fully penetrant maternal effect embryonic lethality, and must be maintained with genetic balancers that require careful monitoring and labor intensive strategies to obtain large populations. In this study, we identified a strong, but viable skn-1 hypomorphic allele skn-1(zj15) from a genetic screen for suppressors of wdr-23, a direct regulator of the transcription factor. skn-1(zj15) is a point mutation in an intron that causes mis-splicing of a fraction of mRNA, and strongly reduces wildtype mRNA levels of the two long skn-1a/c variants. The skn-1(zj15) allele reduces detoxification gene expression and stress resistance to levels comparable to skn-1 RNAi, but, unlike RNAi, it is not restricted from some tissues. We also show that skn-1(zj15) is epistatic to canonical upstream regulators, demonstrating its utility for genetic analysis of skn-1 function and regulation in cases where large numbers of worms are needed, a balancer is problematic, diet is varied, or RNAi cannot be used.
Keywords: genetic screen; mutant; resource.
Copyright © 2016 Tang et al.
Figures


Similar articles
-
Characterization of skn-1/wdr-23 phenotypes in Caenorhabditis elegans; pleiotrophy, aging, glutathione, and interactions with other longevity pathways.Mech Ageing Dev. 2015 Jul;149:88-98. doi: 10.1016/j.mad.2015.06.001. Epub 2015 Jun 6. Mech Ageing Dev. 2015. PMID: 26056713
-
Depletion of a nucleolar protein activates xenobiotic detoxification genes in Caenorhabditis elegans via Nrf /SKN-1 and p53/CEP-1.Free Radic Biol Med. 2012 Mar 1;52(5):937-50. doi: 10.1016/j.freeradbiomed.2011.12.009. Epub 2011 Dec 23. Free Radic Biol Med. 2012. PMID: 22240150
-
Caenorhabditis elegans OSM-11 signaling regulates SKN-1/Nrf during embryonic development and adult longevity and stress response.Dev Biol. 2015 Apr 1;400(1):118-31. doi: 10.1016/j.ydbio.2015.01.021. Epub 2015 Jan 29. Dev Biol. 2015. PMID: 25637691
-
Unique structure and regulation of the nematode detoxification gene regulator, SKN-1: implications to understanding and controlling drug resistance.Drug Metab Rev. 2012 Aug;44(3):209-23. doi: 10.3109/03602532.2012.684799. Epub 2012 Jun 4. Drug Metab Rev. 2012. PMID: 22656429 Free PMC article. Review.
-
A high throughput screen for inhibitors of nematode detoxification genes.2013 Apr 11 [updated 2014 Jan 13]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. 2013 Apr 11 [updated 2014 Jan 13]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. PMID: 25299039 Free Books & Documents. Review.
Cited by
-
Distinct designer diamines promote mitophagy, and thereby enhance healthspan in C. elegans and protect human cells against oxidative damage.Autophagy. 2023 Feb;19(2):474-504. doi: 10.1080/15548627.2022.2078069. Epub 2022 Jun 1. Autophagy. 2023. PMID: 35579620 Free PMC article.
-
The transcription factor SKN-1 and detoxification gene ugt-22 alter albendazole efficacy in Caenorhabditis elegans.Int J Parasitol Drugs Drug Resist. 2018 Aug;8(2):312-319. doi: 10.1016/j.ijpddr.2018.04.006. Epub 2018 Apr 25. Int J Parasitol Drugs Drug Resist. 2018. PMID: 29793058 Free PMC article.
-
Differential impacts of ribosomal protein haploinsufficiency on mitochondrial function.J Cell Biol. 2025 Mar 3;224(3):e202404084. doi: 10.1083/jcb.202404084. Epub 2025 Jan 9. J Cell Biol. 2025. PMID: 39786340
-
An NRF2 Perspective on Stem Cells and Ageing.Front Aging. 2021 Jun 15;2:690686. doi: 10.3389/fragi.2021.690686. eCollection 2021. Front Aging. 2021. PMID: 36213179 Free PMC article. Review.
-
Insulin receptor substrate family member IST-1 regulates the development of Caenorhabditis elegans age-1 and aap-1 mutants.MicroPubl Biol. 2025 Jun 25;2025:10.17912/micropub.biology.001580. doi: 10.17912/micropub.biology.001580. eCollection 2025. MicroPubl Biol. 2025. PMID: 40642152 Free PMC article.
References
-
- Asikainen S., Vartiainen S., Lakso M., Nass R., Wong G., 2005. Selective sensitivity of Caenorhabditis elegans neurons to RNA interference. Neuroreport 16(18): 1995–1999. - PubMed
-
- Bishop N. A., Guarente L., 2007. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447(7144): 545–549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
