Analysis of the mitochondrial maxicircle of Trypanosoma lewisi, a neglected human pathogen
- PMID: 26715306
- PMCID: PMC4696184
- DOI: 10.1186/s13071-015-1281-8
Analysis of the mitochondrial maxicircle of Trypanosoma lewisi, a neglected human pathogen
Erratum in
-
Erratum to: Analysis of the mitochondrial maxicircle of Trypanosoma lewisi, a neglected human pathogen.Parasit Vectors. 2016 Jan 13;9:15. doi: 10.1186/s13071-016-1297-8. Parasit Vectors. 2016. PMID: 26762513 Free PMC article. No abstract available.
Abstract
Background: The haemoflagellate Trypanosoma lewisi is a kinetoplastid parasite which, as it has been recently reported to cause human disease, deserves increased attention. Characteristic features of all kinetoplastid flagellates are a uniquely structured mitochondrial DNA or kinetoplast, comprised of a network of catenated DNA circles, and RNA editing of mitochondrial transcripts. The aim of this study was to describe the kinetoplast DNA of T. lewisi.
Methods/results: In this study, purified kinetoplast DNA from T. lewisi was sequenced using high-throughput sequencing in combination with sequencing of PCR amplicons. This allowed the assembly of the T. lewisi kinetoplast maxicircle DNA, which is a homologue of the mitochondrial genome in other eukaryotes. The assembly of 23,745 bp comprises the non-coding and coding regions. Comparative analysis of the maxicircle sequence of T. lewisi with Trypanosoma cruzi, Trypanosoma rangeli, Trypanosoma brucei and Leishmania tarentolae revealed that it shares 78%, 77%, 74% and 66% sequence identity with these parasites, respectively. The high GC content in at least 9 maxicircle genes of T. lewisi (ATPase6; NADH dehydrogenase subunits ND3, ND7, ND8 and ND9; G-rich regions GR3 and GR4; cytochrome oxidase subunit COIII and ribosomal protein RPS12) implies that their products may be extensively edited. A detailed analysis of the non-coding region revealed that it contains numerous repeat motifs and palindromes.
Conclusions: We have sequenced and comprehensively annotated the kinetoplast maxicircle of T. lewisi. Our analysis reveals that T. lewisi is closely related to T. cruzi and T. brucei, and may share similar RNA editing patterns with them rather than with L. tarentolae. These findings provide novel insight into the biological features of this emerging human pathogen.
Figures
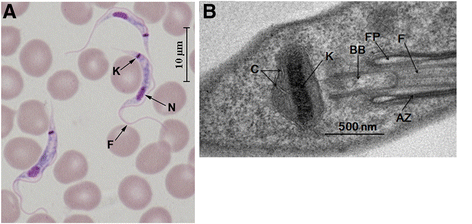



Similar articles
-
Novel insertions in the mitochondrial maxicircle of Trypanosoma musculi, a mouse trypanosome.Parasitology. 2022 Oct;149(12):1546-1555. doi: 10.1017/S0031182022001019. Epub 2022 Aug 4. Parasitology. 2022. PMID: 35924587 Free PMC article.
-
Trypanosoma cruzi mitochondrial maxicircles display species- and strain-specific variation and a conserved element in the non-coding region.BMC Genomics. 2006 Mar 22;7:60. doi: 10.1186/1471-2164-7-60. BMC Genomics. 2006. PMID: 16553959 Free PMC article.
-
The kinetoplast DNA of the Australian trypanosome, Trypanosoma copemani, shares features with Trypanosoma cruzi and Trypanosoma lewisi.Int J Parasitol. 2018 Aug;48(9-10):691-700. doi: 10.1016/j.ijpara.2018.02.006. Epub 2018 May 17. Int J Parasitol. 2018. PMID: 29778329
-
Mitochondrial DNA Structure in Trypanosoma cruzi.Pathogens. 2025 Jan 14;14(1):73. doi: 10.3390/pathogens14010073. Pathogens. 2025. PMID: 39861034 Free PMC article. Review.
-
Kinetoplast DNA in trypanosomid flagellates.Int Rev Cytol. 1986;99:119-79. doi: 10.1016/s0074-7696(08)61426-6. Int Rev Cytol. 1986. PMID: 3082787 Review. No abstract available.
Cited by
-
Leishmania Mitochondrial Genomes: Maxicircle Structure and Heterogeneity of Minicircles.Genes (Basel). 2019 Sep 26;10(10):758. doi: 10.3390/genes10100758. Genes (Basel). 2019. PMID: 31561572 Free PMC article.
-
Cell cycle and cleavage events during in vitro cultivation of bloodstream forms of Trypanosoma lewisi, a zoonotic pathogen.Cell Cycle. 2019 Mar;18(5):552-567. doi: 10.1080/15384101.2019.1577651. Epub 2019 Feb 20. Cell Cycle. 2019. PMID: 30712435 Free PMC article.
-
Erratum to: Analysis of the mitochondrial maxicircle of Trypanosoma lewisi, a neglected human pathogen.Parasit Vectors. 2016 Jan 13;9:15. doi: 10.1186/s13071-016-1297-8. Parasit Vectors. 2016. PMID: 26762513 Free PMC article. No abstract available.
-
Reproduction in Trypanosomatids: Past and Present.Biology (Basel). 2021 May 27;10(6):471. doi: 10.3390/biology10060471. Biology (Basel). 2021. PMID: 34071741 Free PMC article. Review.
-
Novel organization of mitochondrial minicircles and guide RNAs in the zoonotic pathogen Trypanosoma lewisi.Nucleic Acids Res. 2020 Sep 25;48(17):9747-9761. doi: 10.1093/nar/gkaa700. Nucleic Acids Res. 2020. PMID: 32853372 Free PMC article.
References
-
- Hoare CA. The trypanosomes of mammals: A zoological monograph. Oxford: Blackwell Scientific Publications; 1972.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

