Enhanced production of recombinant proteins with Corynebacterium glutamicum by deletion of insertion sequences (IS elements)
- PMID: 26715464
- PMCID: PMC4696348
- DOI: 10.1186/s12934-015-0401-7
Enhanced production of recombinant proteins with Corynebacterium glutamicum by deletion of insertion sequences (IS elements)
Abstract
Background: In most bacteria, various jumping genetic elements including insertion sequences elements (IS elements) cause a variety of genetic rearrangements resulting in harmful effects such as genome and recombinant plasmid instability. The genetic stability of a plasmid in a host is critical for high-level production of recombinant proteins, and in this regard, the development of an IS element-free strain could be a useful strategy for the enhanced production of recombinant proteins. Corynebacterium glutamicum, which is a workhorse in the industrial-scale production of various biomolecules including recombinant proteins, also has several IS elements, and it is necessary to identify the critical IS elements and to develop IS element deleted strain.
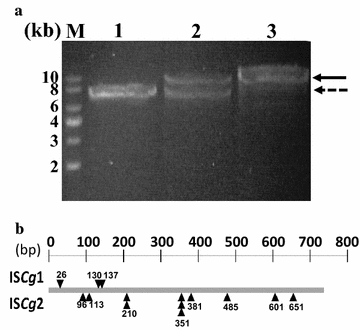
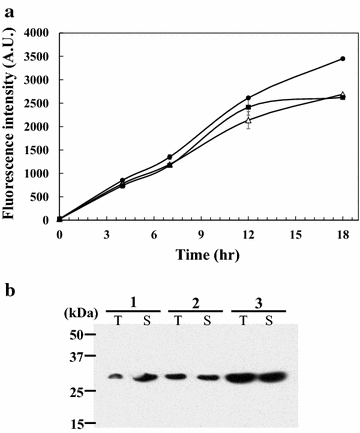
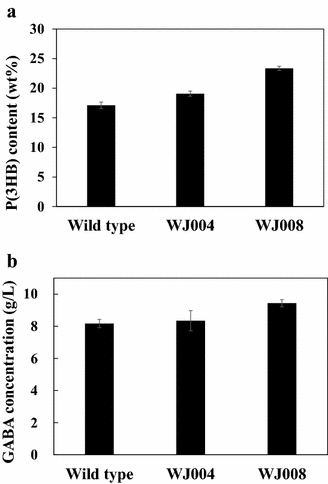
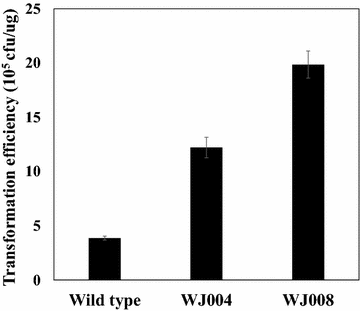
Results: From the cultivation of C. glutamicum harboring a plasmid for green fluorescent protein (GFP) gene expression, non-fluorescent clones were isolated by FACS (fluorescent activated cell sorting). All the isolated clones had insertions of IS elements in the GFP coding region, and two major IS elements (ISCg1 and ISCg2 families) were identified. By co-cultivating cells harboring either the isolated IS element-inserted plasmid or intact plasmid, it was clearly confirmed that cells harboring the IS element-inserted plasmids became dominant during the cultivation due to their growth advantage over cells containing intact plasmids, which can cause a significant reduction in recombinant protein production during cultivation. To minimize the harmful effects of IS elements on the expression of heterologous genes in C. glutamicum, two IS element free C. glutamicum strains were developed in which each major IS element was deleted, and enhanced productivity in the engineered C. glutamicum strain was successfully demonstrated with three models: GFP, poly(3-hydroxybutyrate) [P(3HB)] and γ-aminobutyrate (GABA).
Conclusions: Our findings clearly indicate that the hopping of IS elements could be detrimental to the production of recombinant proteins in C. glutamicum, emphasizing the importance of developing IS element free host strains.
Figures


Similar articles
-
Overexpression of ppc or deletion of mdh for improving production of γ-aminobutyric acid in recombinant Corynebacterium glutamicum.World J Microbiol Biotechnol. 2017 Jun;33(6):122. doi: 10.1007/s11274-017-2289-3. Epub 2017 May 22. World J Microbiol Biotechnol. 2017. PMID: 28534111
-
Enhancing recombinant protein production with an Escherichia coli host strain lacking insertion sequences.Appl Microbiol Biotechnol. 2014 Aug;98(15):6701-13. doi: 10.1007/s00253-014-5739-y. Epub 2014 Apr 22. Appl Microbiol Biotechnol. 2014. PMID: 24752842
-
Development of a high-copy-number plasmid via adaptive laboratory evolution of Corynebacterium glutamicum.Appl Microbiol Biotechnol. 2018 Jan;102(2):873-883. doi: 10.1007/s00253-017-8653-2. Epub 2017 Nov 25. Appl Microbiol Biotechnol. 2018. PMID: 29177939
-
Expression of recombinant protein using Corynebacterium Glutamicum: progress, challenges and applications.Crit Rev Biotechnol. 2016 Aug;36(4):652-64. doi: 10.3109/07388551.2015.1004519. Epub 2015 Feb 25. Crit Rev Biotechnol. 2016. PMID: 25714007 Review.
-
Tools for genetic manipulations in Corynebacterium glutamicum and their applications.Appl Microbiol Biotechnol. 2011 Jun;90(5):1641-54. doi: 10.1007/s00253-011-3272-9. Epub 2011 Apr 26. Appl Microbiol Biotechnol. 2011. PMID: 21519933 Review.
Cited by
-
The future of self-selecting and stable fermentations.J Ind Microbiol Biotechnol. 2020 Nov;47(11):993-1004. doi: 10.1007/s10295-020-02325-0. Epub 2020 Nov 2. J Ind Microbiol Biotechnol. 2020. PMID: 33136197 Free PMC article. Review.
-
Tailoring Corynebacterium glutamicum for Sustainable Biomanufacturing: From Traditional to Cutting-Edge Technologies.Mol Biotechnol. 2025 Jun 10. doi: 10.1007/s12033-025-01447-z. Online ahead of print. Mol Biotechnol. 2025. PMID: 40493161 Review.
-
Frequency, composition and mobility of Escherichia coli-derived transposable elements in holdings of plasmid repositories.Microb Biotechnol. 2022 Feb;15(2):455-468. doi: 10.1111/1751-7915.13962. Epub 2021 Dec 7. Microb Biotechnol. 2022. PMID: 34875147 Free PMC article.
-
Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli.Nucleic Acids Res. 2016 Sep 6;44(15):7109-19. doi: 10.1093/nar/gkw647. Epub 2016 Jul 18. Nucleic Acids Res. 2016. PMID: 27431326 Free PMC article.
-
Construction of an IS-Free Corynebacterium glutamicum ATCC 13 032 Chassis Strain and Random Mutagenesis Using the Endogenous ISCg1 Transposase.Front Bioeng Biotechnol. 2021 Dec 15;9:751334. doi: 10.3389/fbioe.2021.751334. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34976962 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

