Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG
- PMID: 26715750
- PMCID: PMC4720321
- DOI: 10.1073/pnas.1519265113
Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG
Abstract
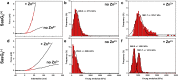
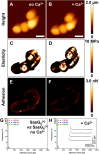
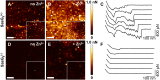
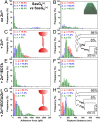
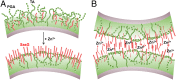
Staphylococcus aureus surface protein SasG promotes cell-cell adhesion during the accumulation phase of biofilm formation, but the molecular basis of this interaction remains poorly understood. Here, we unravel the mechanical properties of SasG on the surface of living bacteria, that is, in its native cellular environment. Nanoscale multiparametric imaging of living bacteria reveals that Zn(2+) strongly increases cell wall rigidity and activates the adhesive function of SasG. Single-cell force measurements show that SasG mediates cell-cell adhesion via specific Zn(2+)-dependent homophilic bonds between β-sheet-rich G5-E domains on neighboring cells. The force required to unfold individual domains is remarkably strong, up to ∼500 pN, thus explaining how SasG can withstand physiological shear forces. We also observe that SasG forms homophilic bonds with the structurally related accumulation-associated protein of Staphylococcus epidermidis, suggesting the possibility of multispecies biofilms during host colonization and infection. Collectively, our findings support a model in which zinc plays a dual role in activating cell-cell adhesion: adsorption of zinc ions to the bacterial cell surface increases cell wall cohesion and favors the projection of elongated SasG proteins away from the cell surface, thereby enabling zinc-dependent homophilic bonds between opposing cells. This work demonstrates an unexpected relationship between mechanics and adhesion in a staphylococcal surface protein, which may represent a general mechanism among bacterial pathogens for activating cell association.
Keywords: SasG; Staphylococcus aureus; adhesion; atomic force microscopy; biofilms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284(5418):1318–1322. - PubMed
-
- Hogan S, Stevens NT, Humphreys H, O’Gara JP, O’Neill E. Current and future approaches to the prevention and treatment of staphylococcal medical device-related infections. Curr Pharm Des. 2015;21(1):100–113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

