Overexpression of miR-199a-5p decreases esophageal cancer cell proliferation through repression of mitogen-activated protein kinase kinase kinase-11 (MAP3K11)
- PMID: 26717044
- PMCID: PMC4891002
- DOI: 10.18632/oncotarget.6752
Overexpression of miR-199a-5p decreases esophageal cancer cell proliferation through repression of mitogen-activated protein kinase kinase kinase-11 (MAP3K11)
Abstract
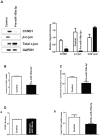
Studies examining the oncogenic or tumor suppressive functions of dysregulated microRNAs (miRs) in cancer cells may also identify novel miR targets, which can themselves serve as therapeutic targets. Using array analysis, we have previously determined that miR-199a-5p was the most downregulated miR in two esophageal cancer cell lines compared to esophageal epithelial cells. MiR-199a-5p is predicted to bind mitogen-activated protein kinase kinase kinase 11 (MAP3K11) mRNA with high affinity. In this study, we observed that MAP3K11 is markedly overexpressed in esophageal cancer cell lines. Forced expression of miR-199a-5p in these cells leads to a decrease in the mRNA and protein levels of MAP3K11, due to decreased MAP3K11 mRNA stability. A direct binding interaction between miR-199a-5p and MAP3K11 mRNA is demonstrated using biotin pull-down assays and heterologous luciferase reporter constructs and confirmed by mutational analysis. Finally, forced expression of miR-199a-5p decreases proliferation of esophageal cancer cells by inducing G2/M arrest. This effect is mediated, in part, by decreased transcription of cyclin D1, due to reduced MAP3K11-mediated phosphorylation of c-Jun. These findings suggest that miR-199a-5p acts as a tumor suppressor in esophageal cancer cells and that its downregulation contributes to enhanced cellular proliferation by targeting MAP3K11.
Keywords: MAP3K11; esophageal cancer; mRNA stability; miR-199a-5p; proliferation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:359–386. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5. - PubMed
-
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. - PubMed
-
- Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

