Antibody diversification caused by disrupted mismatch repair and promiscuous DNA polymerases
- PMID: 26719140
- PMCID: PMC4740194
- DOI: 10.1016/j.dnarep.2015.11.011
Antibody diversification caused by disrupted mismatch repair and promiscuous DNA polymerases
Abstract
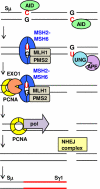
The enzyme activation-induced deaminase (AID) targets the immunoglobulin loci in activated B cells and creates DNA mutations in the antigen-binding variable region and DNA breaks in the switch region through processes known, respectively, as somatic hypermutation and class switch recombination. AID deaminates cytosine to uracil in DNA to create a U:G mismatch. During somatic hypermutation, the MutSα complex binds to the mismatch, and the error-prone DNA polymerase η generates mutations at A and T bases. During class switch recombination, both MutSα and MutLα complexes bind to the mismatch, resulting in double-strand break formation and end-joining. This review is centered on the mechanisms of how the MMR pathway is commandeered by B cells to generate antibody diversity.
Keywords: Activation-induced deaminase; Class switch recombination; DNA polymerase η; Mismatch repair; Somatic hypermutation.
Published by Elsevier B.V.
Figures

References
-
- Subrahmanyam R, Sen R. RAGs' eye view of the immunoglobulin heavy chain gene locus. Semin Immunol. 2010;22:337–345. - PubMed
-
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. - PubMed
-
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
-
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

