Structure-Guided Redesign Increases the Propensity of HIV Env To Generate Highly Stable Soluble Trimers
- PMID: 26719252
- PMCID: PMC4810649
- DOI: 10.1128/JVI.02652-15
Structure-Guided Redesign Increases the Propensity of HIV Env To Generate Highly Stable Soluble Trimers
Abstract
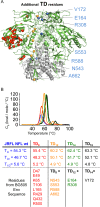
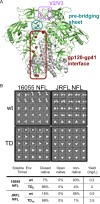
Due to high viral diversity, an effective HIV-1 vaccine will likely require Envs derived from multiple subtypes to generate broadly neutralizing antibodies (bNAbs). Soluble Env mimics, like the native flexibly linked (NFL) and SOSIP trimers, derived from the subtype A BG505 Env, form homogeneous, stable native-like trimers. However, other Env sequences, such as JRFL and 16055 from subtypes B and C, do so to a lesser degree. The high-resolution BG505 SOSIP crystal structures permit the identification and redesign of Env elements involved in trimer stability. Here, we identified structure trimer-derived (TD) residues that increased the propensity of the subtype B JRFL and subtype C 16055 Env sequences to form well-ordered, homogenous, and highly stable soluble trimers. The generation of these spike mimics no longer required antibody-based selection, positive or negative. Using the redesigned subtype B and C trimer representatives as respective foundations, we further stabilized the NFL TD trimers by engineering an intraprotomer disulfide linkage in the prebridging sheet, I201C-A433C (CC), that locks the gp120 in the receptor nontriggered state. We demonstrated that this disulfide pair prevented CD4 induced-conformational rearrangements in NFL trimers derived from the prototypic subtype A, B, and C representatives. Coupling the TD-based design with the engineered disulfide linkage, CC, increased the propensity of Env to form soluble highly stable spike mimics that are resistant to CD4-induced changes. These advances will allow testing of the hypothesis that such stabilized immunogens will more efficiently elicit neutralizing antibodies in small-animal models and primates.
Importance: HIV-1 displays unprecedented global diversity circulating in the human population. Since the envelope glycoprotein (Env) is the target of neutralizing antibodies, Env-based vaccine candidates that address such diversity are needed. Soluble well-ordered Env mimics, typified by NFL and SOSIP trimers, are attractive vaccine candidates. However, the current designs do not allow most Envs to form well-ordered trimers. Here, we made design modifications to increase the propensity of representatives from two of the major HIV subtypes to form highly stable trimers. This approach should be applicable to other viral Envs, permitting the generation of a repertoire of homogeneous, highly stable trimers. The availability of such an array will allow us to assess if sequential or cocktail immune strategies can overcome some of the vaccine challenges presented by HIV diversity.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Gao F, Weaver EA, Lu Z, Li Y, Liao HX, Ma B, Alam SM, Scearce RM, Sutherland LL, Yu JS, Decker JM, Shaw GM, Montefiori DC, Korber BT, Hahn BH, Haynes BF. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol 79:1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. - DOI - PMC - PubMed
-
- Forsell MN, Li Y, Sundback M, Svehla K, Liljestrom P, Mascola JR, Wyatt R, Karlsson Hedestam GB. 2005. Biochemical and immunogenic characterization of soluble human immunodeficiency virus type 1 envelope glycoprotein trimers expressed by Semliki Forest virus. J Virol 79:10902–10914. doi: 10.1128/JVI.79.17.10902-10914.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

