Polyprotein-Driven Formation of Two Interdependent Sets of Complexes Supporting Hepatitis C Virus Genome Replication
- PMID: 26719260
- PMCID: PMC4810661
- DOI: 10.1128/JVI.01931-15
Polyprotein-Driven Formation of Two Interdependent Sets of Complexes Supporting Hepatitis C Virus Genome Replication
Abstract
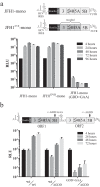
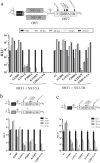
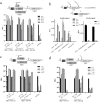
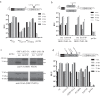
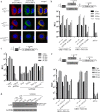
Hepatitis C virus (HCV) requires proteins from the NS3-NS5B polyprotein to create a replicase unit for replication of its genome. The replicase proteins form membranous compartments in cells to facilitate replication, but little is known about their functional organization within these structures. We recently reported on intragenomic replicons, bicistronic viral transcripts expressing an authentic replicase from open reading frame 2 (ORF2) and a second duplicate nonstructural (NS) polyprotein from ORF1. Using these constructs and other methods, we have assessed the polyprotein requirements for rescue of different lethal point mutations across NS3-5B. Mutations readily tractable to rescue broadly fell into two groupings: those requiring expression of a minimum NS3-5A and those requiring expression of a minimum NS3-5B polyprotein. A cis-acting mutation that blocked NS3 helicase activity, T1299A, was tolerated when introduced into either ORF within the intragenomic replicon, but unlike many other mutations required the other ORF to express a functional NS3-5B. Three mutations were identified as more refractile to rescue: one that blocked cleavage of the NS4B5A boundary (S1977P), another in the NS3 helicase (K1240N), and a third in NS4A (V1665G). Introduced into ORF1, these exhibited a dominant negative phenotype, but with K1240N inhibiting replication as a minimum NS3-5A polyprotein whereas V1665G and S1977P only impaired replication as a NS3-5B polyprotein. Furthermore, an S1977P-mutated NS3-5A polyprotein complemented other defects shown to be dependent on NS3-5A for rescue. Overall, our findings suggest the existence of two interdependent sets of protein complexes supporting RNA replication, distinguishable by the minimum polyprotein requirement needed for their formation.
Importance: Positive-strand RNA viruses reshape the intracellular membranes of cells to form a compartment within which to replicate their genome, but little is known about the functional organization of viral proteins within this structure. We have complemented protein-encoded defects in HCV by constructing subgenomic HCV transcripts capable of simultaneously expressing both a mutated and functional polyprotein precursor needed for RNA genome replication (intragenomic replicons). Our results reveal that HCV relies on two interdependent sets of protein complexes to support viral replication. They also show that the intragenomic replicon offers a unique way to study replication complex assembly, as it enables improved composite polyprotein complex formation compared to traditional trans-complementation systems. Finally, the differential behavior of distinct NS3 helicase knockout mutations hints that certain conformations of this enzyme might be particularly deleterious for replication.
Copyright © 2016 Gomes et al.
Figures



Similar articles
-
Genetic complementation of hepatitis C virus nonstructural protein functions associated with replication exhibits requirements that differ from those for virion assembly.J Virol. 2014 Mar;88(5):2748-62. doi: 10.1128/JVI.03588-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352463 Free PMC article.
-
NS5A Domain 1 and Polyprotein Cleavage Kinetics Are Critical for Induction of Double-Membrane Vesicles Associated with Hepatitis C Virus Replication.mBio. 2015 Jul 7;6(4):e00759. doi: 10.1128/mBio.00759-15. mBio. 2015. PMID: 26152585 Free PMC article.
-
Hepatitis C virus RNA replication depends on specific cis- and trans-acting activities of viral nonstructural proteins.PLoS Pathog. 2015 Apr 13;11(4):e1004817. doi: 10.1371/journal.ppat.1004817. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25875808 Free PMC article.
-
Molecular virology of hepatitis C virus: an update with respect to potential antiviral targets.Antivir Ther. 1998;3(Suppl 3):71-81. Antivir Ther. 1998. PMID: 10726057 Review.
-
Molecular virology of the hepatitis C virus.J Hepatol. 1999;31 Suppl 1:47-53. doi: 10.1016/s0168-8278(99)80374-2. J Hepatol. 1999. PMID: 10622560 Review.
Cited by
-
Positive strand RNA viruses differ in the constraints they place on the folding of their negative strand.RNA. 2022 Oct;28(10):1359-1376. doi: 10.1261/rna.079125.122. Epub 2022 Aug 2. RNA. 2022. PMID: 35918125 Free PMC article.
-
Characterization of a multipurpose NS3 surface patch coordinating HCV replicase assembly and virion morphogenesis.PLoS Pathog. 2022 Oct 10;18(10):e1010895. doi: 10.1371/journal.ppat.1010895. eCollection 2022 Oct. PLoS Pathog. 2022. PMID: 36215335 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

