Immunogenicity of a Prefusion HIV-1 Envelope Trimer in Complex with a Quaternary-Structure-Specific Antibody
- PMID: 26719262
- PMCID: PMC4810637
- DOI: 10.1128/JVI.02380-15
Immunogenicity of a Prefusion HIV-1 Envelope Trimer in Complex with a Quaternary-Structure-Specific Antibody
Abstract
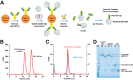
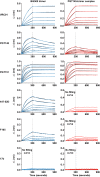
The HIV-1 envelope trimer (Env) is the target of broadly neutralizing antibodies and is being explored as a vaccine candidate to elicit protective antibodies. One of the most promising antigenic and structural mimics of HIV-1 Env is the SOSIP.664-stabilized soluble trimer from the clade A strain BG505, which is preferentially recognized by broadly neutralizing antibodies. Trimer immunization elicits high-titer neutralization of the autologous tier 2 BG505 strain; however, breadth is limited, and substantial interest has focused on understanding and improving trimer immunogenicity. We sought to improve the antigenic specificity of BG505 SOSIP.664 by reducing recognition of the variable loop 3 (V3) region, which elicits only weakly neutralizing antibodies. To stabilize the trimer in its prefusion closed conformation, we complexed trimeric BG505 SOSIP.664 with the antigen-binding fragment (Fab) of PGT145, a broadly neutralizing quaternary-structure-specific antibody. Compared to the ligand-free trimer, the PGT145 Fab-BG505 SOSIP.664 complex displayed increased melting temperature stability and reduced V3 recognition. In guinea pigs, immunization with the PGT145 Fab-BG505 SOSIP.664 complex elicited ∼100-fold lower V3-directed binding and neutralization titers than those obtained with ligand-free BG505 SOSIP.664. Both complexed and ligand-free BG505 SOSIP.664 elicited comparable neutralization of the autologous BG505 virus, and in both cases, BG505 neutralization mapped to the outer domain of gp120 for some guinea pigs. Our results indicate that it is possible to reduce immune recognition of the V3 region of the trimer while maintaining the antigenic profile needed to induce autologous neutralizing antibodies. These data suggest that appropriate modifications of trimer immunogens could further focus the immune response on key neutralization epitopes.
Importance: HIV-1 Env trimers have been proposed as preferred HIV-1 vaccine immunogens. One version, BG505 SOSIP.664, a soluble stabilized trimer, was recently shown to elicit high-titer autologous neutralizing antibodies (NAbs) in rabbits. Here we compared two immunogens: the ligand-free BG505 SOSIP.664 trimer and the same trimer bound to the antigen-binding fragment (Fab) of the PGT145 antibody, a broadly neutralizing antibody which recognizes the trimer at its membrane-distal apex. We hypothesized that the Fab-bound complex would stabilize BG505 SOSIP.664 in its prefusion closed conformation and limit reactivity to weakly neutralizing antibodies targeting the variable loop 3 (V3) region. In guinea pigs, the Fab-complexed trimer induced 100-fold lower responses to the V3 region, while both ligand-free and Fab-complexed trimers elicited similar levels of autologous NAbs. Our findings demonstrate the potential to reduce "off-target" immunogenicity while maintaining the capacity to generate autologous NAbs.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures







References
-
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

