A Cyclin-Dependent Kinase Inhibitor, Dinaciclib, Impairs Homologous Recombination and Sensitizes Multiple Myeloma Cells to PARP Inhibition
- PMID: 26719576
- PMCID: PMC4747838
- DOI: 10.1158/1535-7163.MCT-15-0660
A Cyclin-Dependent Kinase Inhibitor, Dinaciclib, Impairs Homologous Recombination and Sensitizes Multiple Myeloma Cells to PARP Inhibition
Abstract
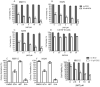
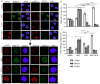
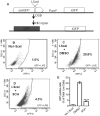
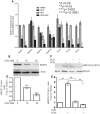
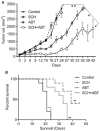
PARP1/2 are required for single-strand break repair, and their inhibition causes DNA replication fork collapse and double-strand break (DSB) formation. These DSBs are primarily repaired via homologous recombination (HR), a high-fidelity repair pathway. Should HR be deficient, DSBs may be repaired via error-prone nonhomologous end-joining mechanisms, or may persist, ultimately resulting in cell death. The combined disruption of PARP and HR activities thus produces synthetic lethality. Multiple myeloma cells are characterized by chromosomal instability and pervasive DNA damage, implicating aberrant DNA repair. Cyclin-dependent kinases (CDK), upstream modulators of HR, are dysregulated in multiple myeloma. Here, we show that a CDK inhibitor, dinaciclib, impairs HR repair and sensitizes multiple myeloma cells to the PARP1/2 inhibitor ABT-888. Dinaciclib abolishes ABT-888-induced BRCA1 and RAD51 foci and potentiates DNA damage, indicated by increased γH2AX foci. Dinaciclib treatment reduces expression of HR repair genes, including Rad51, and blocks BRCA1 phosphorylation, a modification required for HR repair, thus inhibiting HR repair of chromosome DSBs. Cotreatment with dinaciclib and ABT-888 in vitro resulted in synthetic lethality of multiple myeloma cells, but not normal CD19(+) B cells, and slowed growth of multiple myeloma xenografts in SCID mice almost two-fold. These findings support combining dinaciclib with PARP inhibitors for multiple myeloma therapy. Mol Cancer Ther; 15(2); 241-50. ©2015 AACR.
©2015 American Association for Cancer Research.
Conflict of interest statement
Figures
References
-
- Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–48. - PubMed
-
- Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–22. - PubMed
-
- Velangi MR, Matheson EC, Morgan GJ, Jackson GH, Taylor PR, Hall AG, et al. DNA mismatch repair pathway defects in the pathogenesis and evolution of myeloma. Carcinogenesis. 2004;25:1795–803. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

