Derivation of consensus inactivation status for X-linked genes from genome-wide studies
- PMID: 26719789
- PMCID: PMC4696107
- DOI: 10.1186/s13293-015-0053-7
Derivation of consensus inactivation status for X-linked genes from genome-wide studies
Abstract
Background: X chromosome inactivation is the epigenetic silencing of the majority of the genes on one of the X chromosomes in XX therian mammals. In humans, approximately 15 % of genes consistently escape from this inactivation and another 15 % of genes vary between individuals or tissues in whether they are subject to, or escape from, inactivation. Multiple studies have provided inactivation status calls for a large subset of the genes on the X chromosome; however, these studies vary in which genes they were able to make calls for and in some cases which call they give a specific gene.
Methods: This analysis aggregated three published studies that have examined X chromosome inactivation status of genes across the X chromosome, generating consensus calls and identifying discordancies. The impact of expression level and chromosomal location on X chromosome inactivation status was also assessed.
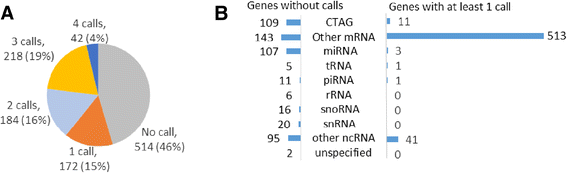
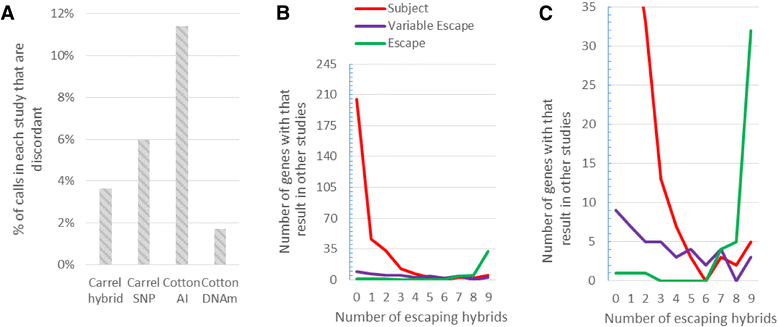
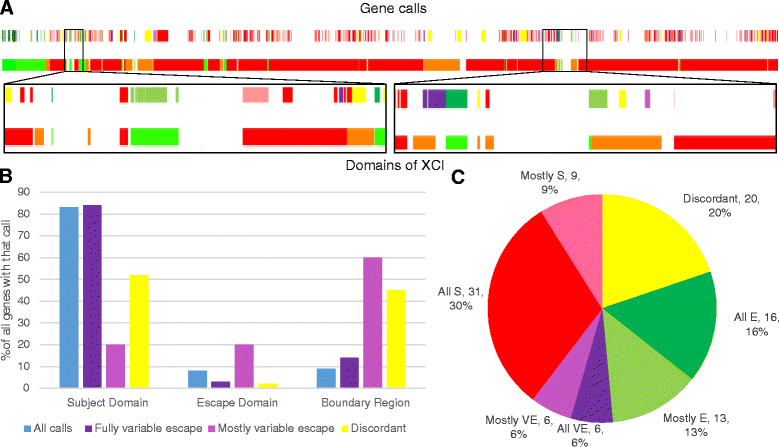
Results: Overall, we assigned a consensus XCI status 639 genes, including 78 % of protein-coding genes expressed outside of the testes, with a lower frequency for non-coding RNA and testis-specific genes. Study-specific discordancies suggest that there may be instability of XCI during cell culture and also highlight study-specific variations in call type. We observe an enrichment of discordant genes at boundaries between genes subject to and escaping from inactivation.
Conclusions: This study has compiled a comprehensive list of X-chromosome inactivation statuses for genes and also discovered some biases which will help guide future studies examining X-chromosome inactivation.
Keywords: Allelic imbalance; DNA methylation; Dosage compensation; Escape from X-chromosome inactivation; Somatic cell hybrids; X-chromosome inactivation.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

