TRIO loss of function is associated with mild intellectual disability and affects dendritic branching and synapse function
- PMID: 26721934
- PMCID: PMC4754042
- DOI: 10.1093/hmg/ddv618
TRIO loss of function is associated with mild intellectual disability and affects dendritic branching and synapse function
Abstract
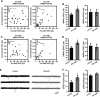
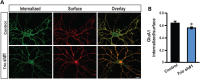
Recently, we marked TRIO for the first time as a candidate gene for intellectual disability (ID). Across diverse vertebrate species, TRIO is a well-conserved Rho GTPase regulator that is highly expressed in the developing brain. However, little is known about the specific events regulated by TRIO during brain development and its clinical impact in humans when mutated. Routine clinical diagnostic testing identified an intragenic de novo deletion of TRIO in a boy with ID. Targeted sequencing of this gene in over 2300 individuals with ID, identified three additional truncating mutations. All index cases had mild to borderline ID combined with behavioral problems consisting of autistic, hyperactive and/or aggressive behavior. Studies in dissociated rat hippocampal neurons demonstrated the enhancement of dendritic formation by suppressing endogenous TRIO, and similarly decreasing endogenous TRIO in organotypic hippocampal brain slices significantly increased synaptic strength by increasing functional synapses. Together, our findings provide new mechanistic insight into how genetic deficits in TRIO can lead to early neuronal network formation by directly affecting both neurite outgrowth and synapse development.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Leonard H., Wen X. (2002) The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment. Retard. Dev. Disabil. Res. Rev., 8, 117–134. - PubMed
-
- Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. (2011) Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet., 12, 745–755. - PubMed
-
- Vissers L.E.L.M., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M. et al. (2010) A de novo paradigm for mental retardation. Nat. Genet., 42, 1109–1112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

