Molecular characterization and phylogenetic analysis of pseudorabies virus variants isolated from Guangdong province of southern China during 2013-2014
- PMID: 26726029
- PMCID: PMC5037305
- DOI: 10.4142/jvs.2016.17.3.369
Molecular characterization and phylogenetic analysis of pseudorabies virus variants isolated from Guangdong province of southern China during 2013-2014
Abstract
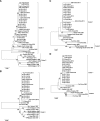
Outbreaks of pseudorabies (PR) have occurred in southern China since late 2011, resulting in significant economic impacts on the swine industry. To identify the cause of PR outbreaks, especially among vaccinated pigs, 11 pseudorabies virus (PRV) field strains were isolated from Guangdong province during 2013-2014. Their major viral genes (gE, TK, gI, PK, gD, 11K, and 28K) were analyzed in this study. Insertions or deletions were observed in gD, gE, gI and PK genes compared with other PRV isolates from all over the world. Furthermore, sequence alignment showed that insertions in gD and gE were unique molecular characteristics of the new prevalent PRV strains in China. Phylogenetic analysis showed that our isolates were clustered in an independent branch together with other strains isolated from China in recent years, and that they showed a closer genetic relationship with earlier isolates from Asia. Our results suggest that these isolates are novel PRV variants with unique molecular signatures.
Keywords: molecular characterization; phylogenetic analysis; pseudorabies virus.
Conflict of interest statement
There is no conflict of interest.
Figures


Similar articles
-
Molecular epidemiology of outbreak-associated pseudorabies virus (PRV) strains in central China.Virus Genes. 2015 Jun;50(3):401-9. doi: 10.1007/s11262-015-1190-0. Epub 2015 Apr 10. Virus Genes. 2015. PMID: 25860998
-
Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China.Vet Microbiol. 2014 Nov 7;174(1-2):107-15. doi: 10.1016/j.vetmic.2014.09.003. Epub 2014 Sep 22. Vet Microbiol. 2014. PMID: 25293398
-
Seroprevalence investigation and genetic analysis of pseudorabies virus within pig populations in Henan province of China during 2018-2019.Infect Genet Evol. 2021 Aug;92:104835. doi: 10.1016/j.meegid.2021.104835. Epub 2021 Mar 31. Infect Genet Evol. 2021. PMID: 33798759
-
Pseudorabies virus in wild swine: a global perspective.Arch Virol. 2011 Oct;156(10):1691-705. doi: 10.1007/s00705-011-1080-2. Epub 2011 Aug 12. Arch Virol. 2011. PMID: 21837416 Review.
-
Current Status and Challenge of Pseudorabies Virus Infection in China.Virol Sin. 2021 Aug;36(4):588-607. doi: 10.1007/s12250-020-00340-0. Epub 2021 Feb 22. Virol Sin. 2021. PMID: 33616892 Free PMC article. Review.
Cited by
-
Recombinant Pseudorabies Virus Usage in Vaccine Development against Swine Infectious Disease.Viruses. 2023 Jan 28;15(2):370. doi: 10.3390/v15020370. Viruses. 2023. PMID: 36851584 Free PMC article. Review.
-
Pseudorabies Virus: From Pathogenesis to Prevention Strategies.Viruses. 2022 Jul 27;14(8):1638. doi: 10.3390/v14081638. Viruses. 2022. PMID: 36016260 Free PMC article. Review.
-
An inactivated gE-deleted pseudorabies vaccine provides complete clinical protection and reduces virus shedding against challenge by a Chinese pseudorabies variant.BMC Vet Res. 2016 Dec 7;12(1):277. doi: 10.1186/s12917-016-0897-z. BMC Vet Res. 2016. PMID: 27923365 Free PMC article.
-
Isolation and Characterization of a Variant Psedorabies Virus HNXY and Construction of rHNXY-∆TK/∆gE.Animals (Basel). 2020 Oct 4;10(10):1804. doi: 10.3390/ani10101804. Animals (Basel). 2020. PMID: 33020441 Free PMC article.
-
Construction and Immunogenicity of a Recombinant Pseudorabies Virus Variant With TK/gI/gE/11k/28k Deletion.Front Vet Sci. 2022 Jan 25;8:797611. doi: 10.3389/fvets.2021.797611. eCollection 2021. Front Vet Sci. 2022. PMID: 35146013 Free PMC article.
References
-
- Eloit M, Fargeaud D, L'Haridon R, Toma B. Identification of the pseudorabies virus glycoprotein gp50 as a major target of neutralizing antibodies. Arch Virol. 1988;99:45–56. - PubMed
-
- Ficińska J, Bieńkowska-Szewczyk K, Jacobs L, Płucienniczak G, Płucienniczak A, Szewczyk B. Characterization of changes in the short unique segment of pseudorabies virus BUK-TK900 (Suivac A) vaccine strain. Arch Virol. 2003;148:1593–1612. - PubMed
-
- Kimman TG, de Wind N, Oei-Lie N, Pol JMA, Berns AJM, Gielkens ALJ. Contribution of single genes within the unique short region of Aujeszk's disease virus (suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J Gen Virol. 1992;73:243–251. - PubMed
-
- Kit S, Kit M, Pirtle EC. Attenuated properties of thymidine kinase-negative deletion mutant of pseudorabies virus. Am J Vet Res. 1985;46:1359–1367. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

