Deciphering the Code of the Cancer Genome: Mechanisms of Chromosome Rearrangement
- PMID: 26726318
- PMCID: PMC4695301
- DOI: 10.1016/j.trecan.2015.10.007
Deciphering the Code of the Cancer Genome: Mechanisms of Chromosome Rearrangement
Abstract
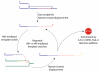
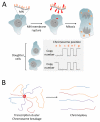
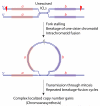
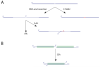
Chromosome rearrangement plays a causal role in tumorigenesis by contributing to the inactivation of tumor suppressor genes, the dysregulated expression or amplification of oncogenes and the generation of novel gene fusions. Chromosome breaks are important intermediates in this process. How, when and where these breaks arise and the specific mechanisms engaged in their repair strongly influence the resulting patterns of chromosome rearrangement. Here, we review recent progress in understanding how certain distinctive features of the cancer genome, including clustered mutagenesis, tandem segmental duplications, complex breakpoints, chromothripsis, chromoplexy and chromoanasynthesis may arise.
Figures


References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends in cell biology. 1999;9:M57–60. - PubMed
-
- Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16:207–220. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
