Rumen bacterial communities can be acclimated faster to high concentrate diets than currently implemented feedlot programs
- PMID: 26726754
- PMCID: PMC4785609
- DOI: 10.1111/jam.13039
Rumen bacterial communities can be acclimated faster to high concentrate diets than currently implemented feedlot programs
Abstract
Aims: Recent studies have demonstrated RAMP, a complete starter feed, to have beneficial effects for animal performance. However, how RAMP may elicit such responses is unknown. To understand if RAMP adaptation results in changes in the rumen bacterial community that can potentially affect animal performance, we investigated the dynamics of rumen bacterial community composition in corn-adapted and RAMP-adapted cattle.
Methods and results: During gradual acclimation of the rumen bacterial communities, we compared the bacterial community dynamics in corn and RAMP-adapted using 16S rRNA gene amplicon sequencing. Significant shifts in bacterial populations across diets were identified. The shift in corn-adapted animals occurred between adaptation step3 and step4, whereas in RAMP-adapted cattle, the shift occurred between step2 and step3. As the adaptation program progressed, the abundance of OTUs associated with family Prevotellaceae and S24-7 changed in corn-adapted animals. In RAMP-adapted animals, OTUs belonging to family Ruminococcaceae and Lachnospiraceae changed in abundance.
Conclusions: Rumen bacteria can be acclimated faster to high concentrate diets, such as RAMP, than traditional adaptation programs and the speed of bacterial community acclimation depends on substrate composition.
Significance and impact of the study: These findings may have implications for beef producers to reduce feedlot costs, as less time adapting animals would result in lower feed costs. However, animal feeding behavior patterns and other factors must be considered.
Keywords: 16S rRNA; RAMP; bacteria; microbial community; rumen microbiology.
© 2016 The Authors published by John Wiley & Sons Ltd on behalf of Society for Applied Microbiology.
Figures
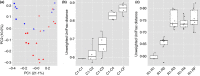
 ), C1; (
), C1; ( ), C2; (
), C2; ( ), C3; (
), C3; ( ), C4; (
), C4; ( ), CF; (
), CF; ( ), R1; (
), R1; ( ), R2; (
), R2; ( ), R3; (
), R3; ( ), R4; (
), R4; ( ), RF.
), RF.


References
-
- Bevans, D.W. , Beauchemin, K.A. , Schwartzkopf‐Genswein, K.S. , McKinnon, J.J. and McAllister, T.A. (2005) Effect of rapid or gradual grain adaptation on subacute acidosis and feed intake by feedlot cattle. J Anim Sci 83, 1116–1132. - PubMed
-
- Buttrey, E.K. , Luebbe, M.K. , Bondurant, R.G. and MacDonald, J.C. (2012) Case Study: grain adaptation of yearling steers to steam‐flaked corn‐based diets using a complete starter feed. Prof Anim Sci 7, 482–488.
-
- Carlisle, E.M. , Poroyko, V. , Caplan, M.S. , Alverdy, J. , Morowitz, M.J. and Liu, D. (2013) Murine gut microbiota and transcriptome are diet dependent. Ann Surg 257, 287–294. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

