Adult mouse cortical cell taxonomy revealed by single cell transcriptomics
- PMID: 26727548
- PMCID: PMC4985242
- DOI: 10.1038/nn.4216
Adult mouse cortical cell taxonomy revealed by single cell transcriptomics
Abstract
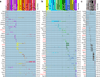
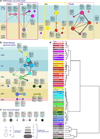
Nervous systems are composed of various cell types, but the extent of cell type diversity is poorly understood. We constructed a cellular taxonomy of one cortical region, primary visual cortex, in adult mice on the basis of single-cell RNA sequencing. We identified 49 transcriptomic cell types, including 23 GABAergic, 19 glutamatergic and 7 non-neuronal types. We also analyzed cell type-specific mRNA processing and characterized genetic access to these transcriptomic types by many transgenic Cre lines. Finally, we found that some of our transcriptomic cell types displayed specific and differential electrophysiological and axon projection properties, thereby confirming that the single-cell transcriptomic signatures can be associated with specific cellular properties.
Figures





Comment in
-
Seq-ing the cortex one neuron at a time.Nat Neurosci. 2016 Feb;19(2):179-81. doi: 10.1038/nn.4230. Nat Neurosci. 2016. PMID: 26814585 No abstract available.
References
Additional references
-
- Dhillon H, et al. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron. 2006;49:191–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

