How type 1 fimbriae help Escherichia coli to evade extracellular antibiotics
- PMID: 26728082
- PMCID: PMC4700443
- DOI: 10.1038/srep18109
How type 1 fimbriae help Escherichia coli to evade extracellular antibiotics
Abstract
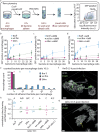
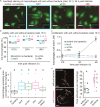
To survive antibiotics, bacteria use two different strategies: counteracting antibiotic effects by expression of resistance genes or evading their effects e.g. by persisting inside host cells. Since bacterial adhesins provide access to the shielded, intracellular niche and the adhesin type 1 fimbriae increases bacterial survival chances inside macrophages, we asked if fimbriae also influenced survival by antibiotic evasion. Combined gentamicin survival assays, flow cytometry, single cell microscopy and kinetic modeling of dose response curves showed that type 1 fimbriae increased the adhesion and internalization by macrophages. This was caused by strongly decreased off-rates and affected the number of intracellular bacteria but not the macrophage viability and morphology. Fimbriae thus promote antibiotic evasion which is particularly relevant in the context of chronic infections.
Figures


Similar articles
-
Differential effects of antibiotics on adhesins of antibiotic resistant strains of Escherichia coli.Scand J Infect Dis Suppl. 1982;33:108-14. Scand J Infect Dis Suppl. 1982. PMID: 6127799
-
Influence of extracellular bactericidal agents on bacteria within macrophages.Infect Immun. 2003 Feb;71(2):1016-9. doi: 10.1128/IAI.71.2.1016-1019.2003. Infect Immun. 2003. PMID: 12540587 Free PMC article.
-
Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic.Nature. 1997 Oct 9;389(6651):636-9. doi: 10.1038/39376. Nature. 1997. PMID: 9335508
-
[Effect of antibiotics on microbial adhesion].Antibiot Khimioter. 1990 Mar;35(3):50-3. Antibiot Khimioter. 1990. PMID: 1972877 Review. Russian. No abstract available.
-
Bacterial structure and its implications in the mechanisms of infection: a short review.Can J Microbiol. 1980 Jun;26(6):643-53. doi: 10.1139/m80-113. Can J Microbiol. 1980. PMID: 6105014 Review. No abstract available.
Cited by
-
MgrB Mutations and Altered Cell Permeability in Colistin Resistance in Klebsiella pneumoniae.Cells. 2022 Sep 26;11(19):2995. doi: 10.3390/cells11192995. Cells. 2022. PMID: 36230959 Free PMC article. Review.
-
Correlation analysis of whole genome sequencing of a pathogenic Escherichia coli strain of Inner Mongolian origin.Sci Rep. 2024 Jul 5;14(1):15494. doi: 10.1038/s41598-024-64256-5. Sci Rep. 2024. PMID: 38969720 Free PMC article.
-
Genomic diversity, pathogenicity and antimicrobial resistance of Escherichia coli isolated from poultry in the southern United States.BMC Microbiol. 2023 Jan 16;23(1):15. doi: 10.1186/s12866-022-02721-9. BMC Microbiol. 2023. PMID: 36647025 Free PMC article.
-
Non-lethal exposure to H2O2 boosts bacterial survival and evolvability against oxidative stress.PLoS Genet. 2020 Mar 12;16(3):e1008649. doi: 10.1371/journal.pgen.1008649. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32163413 Free PMC article.
-
Simulated Gastric Acid Promotes the Horizontal Transfer of Multidrug Resistance Genes across Bacteria in the Gastrointestinal Tract at Elevated pH Levels.Microbiol Spectr. 2023 Jun 15;11(3):e0482022. doi: 10.1128/spectrum.04820-22. Epub 2023 Apr 18. Microbiol Spectr. 2023. PMID: 37070984 Free PMC article.
References
-
- Garcia-del Portillo F. & Finlay B. B. The varied lifestyles of intracellular pathogens within eukaryotic vacuolar compartments. Trends in microbiology 3, 373–380 (1995). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

