Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames
- PMID: 26728094
- PMCID: PMC4728431
- DOI: 10.1038/ncomms10238
Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames
Abstract
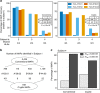
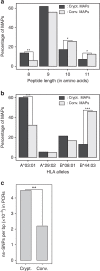
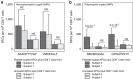
In view of recent reports documenting pervasive translation outside of canonical protein-coding sequences, we wished to determine the proportion of major histocompatibility complex (MHC) class I-associated peptides (MAPs) derived from non-canonical reading frames. Here we perform proteogenomic analyses of MAPs eluted from human B cells using high-throughput mass spectrometry to probe the six-frame translation of the B-cell transcriptome. We report that ∼ 10% of MAPs originate from allegedly noncoding genomic sequences or exonic out-of-frame translation. The biogenesis and properties of these 'cryptic MAPs' differ from those of conventional MAPs. Cryptic MAPs come from very short proteins with atypical C termini, and are coded by transcripts bearing long 3'UTRs enriched in destabilizing elements. Relative to conventional MAPs, cryptic MAPs display different MHC class I-binding preferences and harbour more genomic polymorphisms, some of which are immunogenic. Cryptic MAPs increase the complexity of the MAP repertoire and enhance the scope of CD8 T-cell immunosurveillance.
Figures




References
-
- Alfaro J. A., Sinha A., Kislinger T. & Boutros P. C. Onco-proteogenomics: cancer proteomics joins forces with genomics. Nat. Methods 11, 1107–1113 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

