High-fat-diet induced development of increased fasting glucose levels and impaired response to intraperitoneal glucose challenge in the collaborative cross mouse genetic reference population
- PMID: 26728312
- PMCID: PMC4700737
- DOI: 10.1186/s12863-015-0321-x
High-fat-diet induced development of increased fasting glucose levels and impaired response to intraperitoneal glucose challenge in the collaborative cross mouse genetic reference population
Abstract
Background: The prevalence of Type 2 Diabetes (T2D) mellitus in the past decades, has reached epidemic proportions. Several lines of evidence support the role of genetic variation in the pathogenesis of T2D and insulin resistance. Elucidating these factors could contribute to developing new medical treatments and tools to identify those most at risk. The aim of this study was to characterize the phenotypic response of the Collaborative Cross (CC) mouse genetic resource population to high-fat diet (HFD) induced T2D-like disease to evaluate its suitability for this purpose.
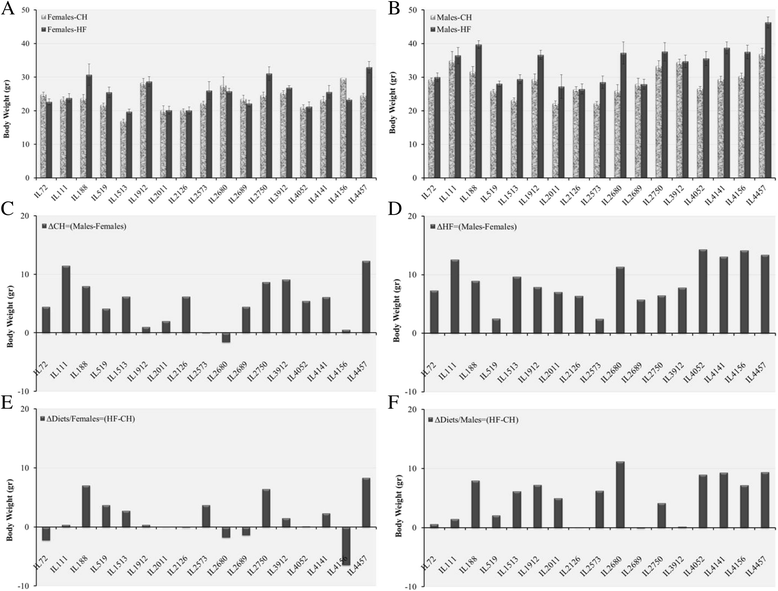
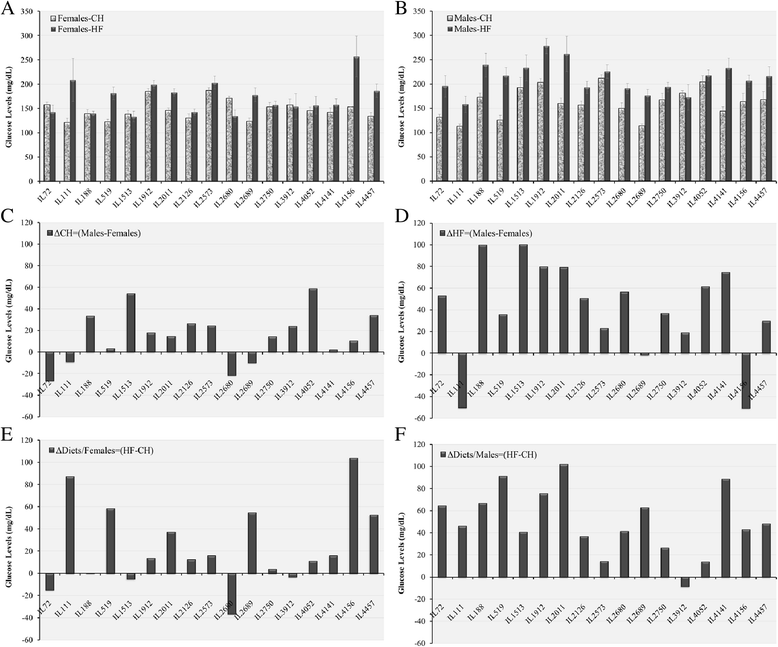
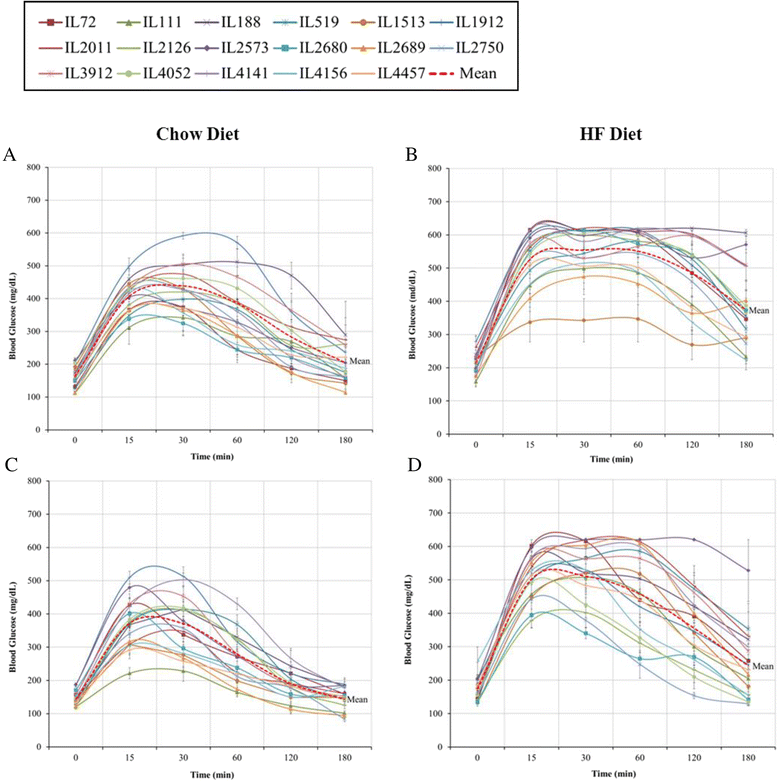
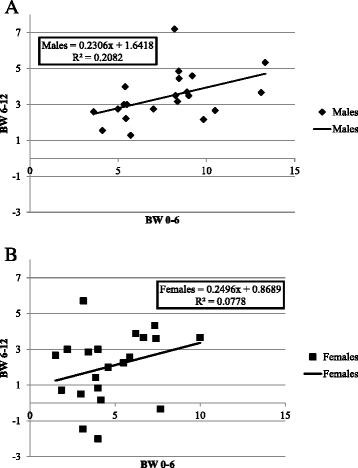
Results: We studied 683 mice of 21 different lines of the CC population. Of these, 265 mice (149 males and 116 females) were challenged by HFD (42% fat); and 384 mice (239 males and 145 females) of 17 of the 21 lines were reared as control group on standard Chow diet (18% fat). Briefly, 8 week old mice were maintained on HFD until 20 weeks of age, and subsequently assessed by intraperitoneal glucose tolerance test (IPGTT). Biweekly body weight (BW), body length (BL), waist circumstance (WC), and body mass index (BMI) were measured. On statistical analysis, trait measurements taken at 20 weeks of age showed significant sex by diet interaction across the different lines and traits. Consequently, males and females were analyzed, separately. Differences among lines were analyzed by ANOVA and shown to be significant (P <0.05), for BW, WC, BMI, fasting blood glucose, and IPGTT-AUC. We use these data to infer broad sense heritability adjusted for number of mice tested in each line; coefficient of genetic variation; genetic correlations between the same trait in the two sexes, and phenotypic correlations between different traits in the same sex.
Conclusions: These results are consistent with the hypothesis that host susceptibility to HFD-induced T2D is a complex trait and controlled by multiple genetic factors and sex, and that the CC population can be a powerful tool for genetic dissection of this trait.
Figures


Similar articles
-
Studying host genetic background effects on multimorbidity of intestinal cancer development, type 2 diabetes and obesity in response to oral bacterial infection and high-fat diet using the collaborative cross (CC) lines.Animal Model Exp Med. 2021 Feb 14;4(1):27-39. doi: 10.1002/ame2.12151. eCollection 2021 Mar. Animal Model Exp Med. 2021. PMID: 33738434 Free PMC article.
-
Efficient protocols and methods for high-throughput utilization of the Collaborative Cross mouse model for dissecting the genetic basis of complex traits.Animal Model Exp Med. 2019 Jul 10;2(3):137-149. doi: 10.1002/ame2.12074. eCollection 2019 Sep. Animal Model Exp Med. 2019. PMID: 31773089 Free PMC article. Review.
-
Intestinal cancer development in response to oral infection with high-fat diet-induced Type 2 diabetes (T2D) in collaborative cross mice under different host genetic background effects.Mamm Genome. 2023 Mar;34(1):56-75. doi: 10.1007/s00335-023-09979-y. Epub 2023 Feb 9. Mamm Genome. 2023. PMID: 36757430
-
Unraveling the Host Genetic Background Effect on Internal Organ Weight Influenced by Obesity and Diabetes Using Collaborative Cross Mice.Int J Mol Sci. 2023 May 3;24(9):8201. doi: 10.3390/ijms24098201. Int J Mol Sci. 2023. PMID: 37175908 Free PMC article.
-
Host Genetics Background Affects Intestinal Cancer Development Associated with High-Fat Diet-Induced Obesity and Type 2 Diabetes.Cells. 2024 Oct 31;13(21):1805. doi: 10.3390/cells13211805. Cells. 2024. PMID: 39513912 Free PMC article.
Cited by
-
The Collaborative Cross mouse model for dissecting genetic susceptibility to infectious diseases.Mamm Genome. 2018 Aug;29(7-8):471-487. doi: 10.1007/s00335-018-9768-1. Epub 2018 Aug 24. Mamm Genome. 2018. PMID: 30143822 Review.
-
The potential of integrating human and mouse discovery platforms to advance our understanding of cardiometabolic diseases.Elife. 2023 Mar 31;12:e86139. doi: 10.7554/eLife.86139. Elife. 2023. PMID: 37000167 Free PMC article.
-
Container-aided integrative QTL and RNA-seq analysis of Collaborative Cross mice supports distinct sex-oriented molecular modes of response in obesity.BMC Genomics. 2020 Nov 3;21(1):761. doi: 10.1186/s12864-020-07173-x. BMC Genomics. 2020. PMID: 33143653 Free PMC article.
-
Studying host genetic background effects on multimorbidity of intestinal cancer development, type 2 diabetes and obesity in response to oral bacterial infection and high-fat diet using the collaborative cross (CC) lines.Animal Model Exp Med. 2021 Feb 14;4(1):27-39. doi: 10.1002/ame2.12151. eCollection 2021 Mar. Animal Model Exp Med. 2021. PMID: 33738434 Free PMC article.
-
Efficient protocols and methods for high-throughput utilization of the Collaborative Cross mouse model for dissecting the genetic basis of complex traits.Animal Model Exp Med. 2019 Jul 10;2(3):137-149. doi: 10.1002/ame2.12074. eCollection 2019 Sep. Animal Model Exp Med. 2019. PMID: 31773089 Free PMC article. Review.
References
-
- National Diabetes Information Clearinghouse. National Diabetes Statistics. NIH Publication. http://www.cdc.gov/diabetes/pubs/statsreport14/national-diabetes-report-.... (2014). Accessed 26 Apr 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

