HIV-1 latency and virus production from unintegrated genomes following direct infection of resting CD4 T cells
- PMID: 26728316
- PMCID: PMC4700562
- DOI: 10.1186/s12977-015-0234-9
HIV-1 latency and virus production from unintegrated genomes following direct infection of resting CD4 T cells
Abstract
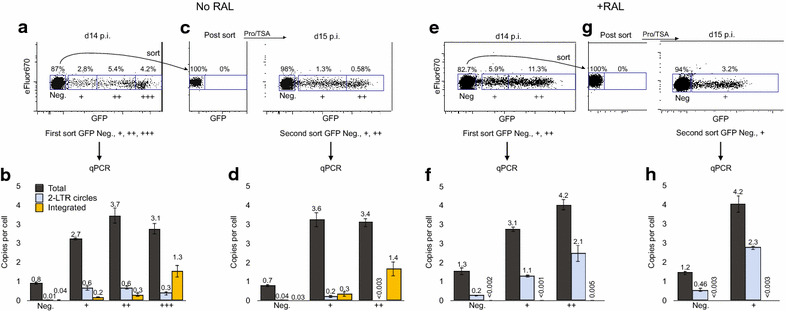
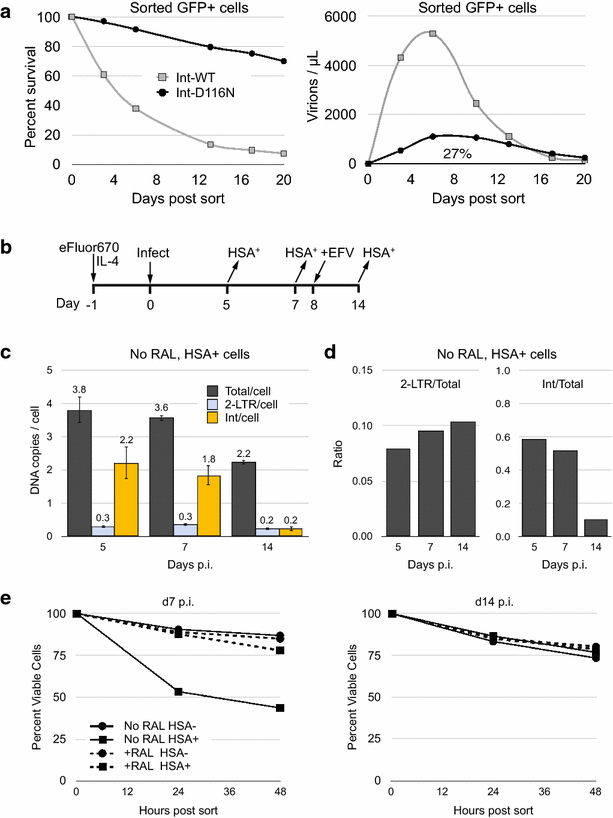
Background: HIV-1 integration is prone to a high rate of failure, resulting in the accumulation of unintegrated viral genomes (uDNA) in vivo and in vitro. uDNA can be transcriptionally active, and circularized uDNA genomes are biochemically stable in non-proliferating cells. Resting, non-proliferating CD4 T cells are prime targets of HIV-1 infection and latently infected resting CD4 T cells are the major barrier to HIV cure. Our prior studies demonstrated that uDNA generates infectious virions when T cell activation follows rather than precedes infection.
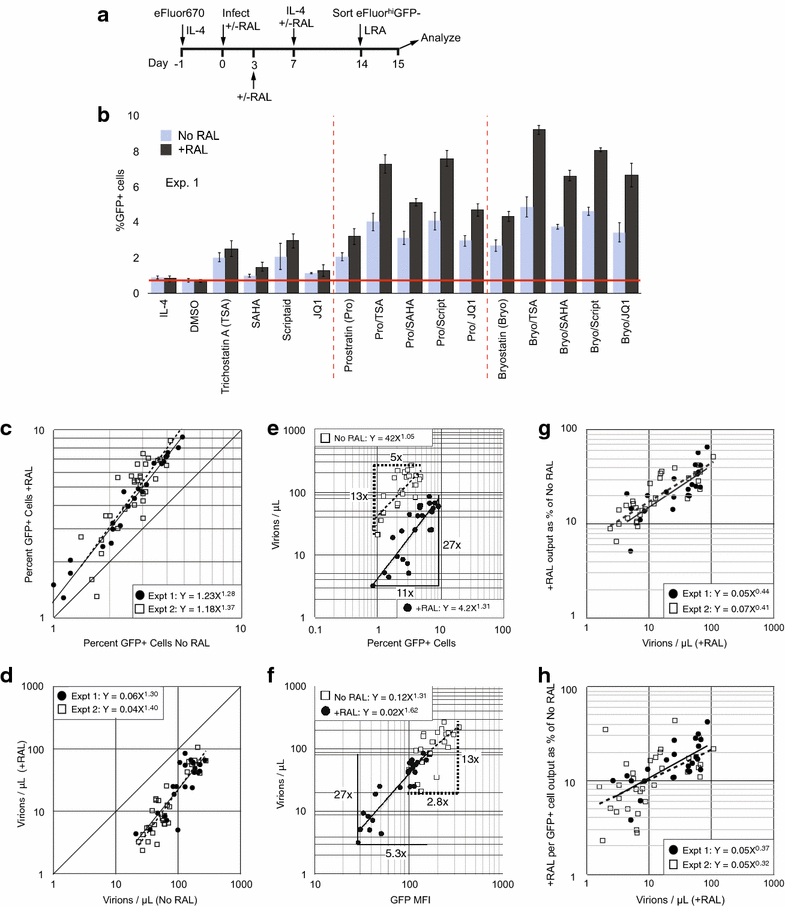
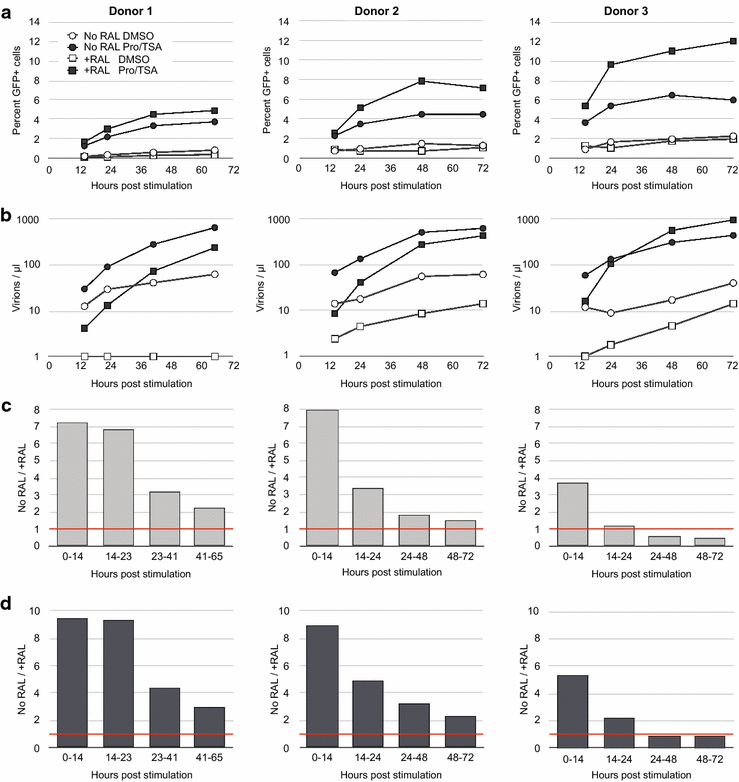
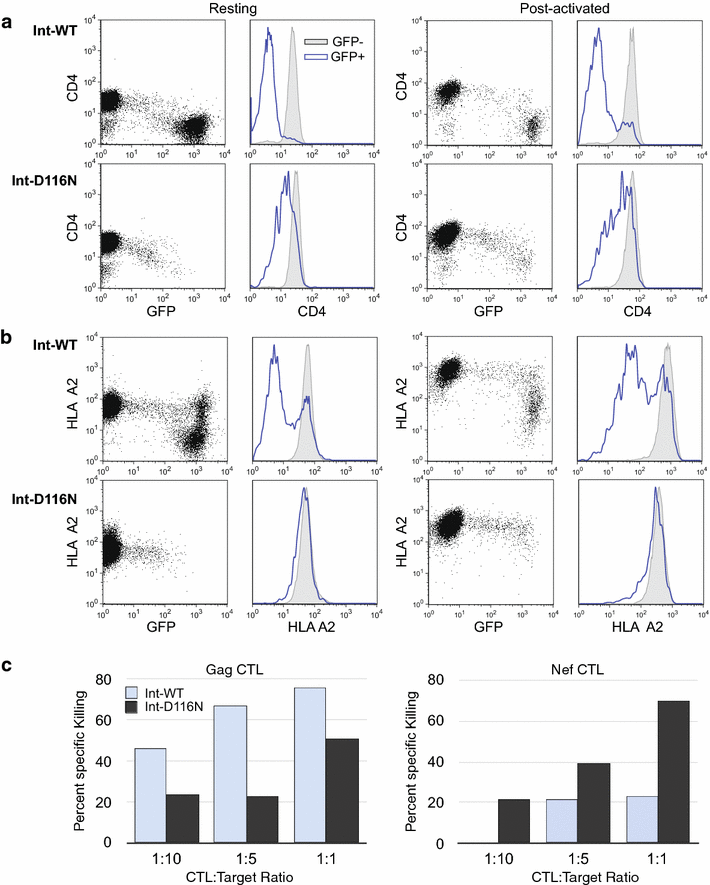
Results: Here, we characterize in primary resting CD4 T cells the dynamics of integrated and unintegrated virus expression, genome persistence and sensitivity to latency reversing agents. Unintegrated HIV-1 was abundant in directly infected resting CD4 T cells. Maximal gene expression from uDNA was delayed compared with integrated HIV-1 and was less toxic, resulting in uDNA enrichment over time relative to integrated proviruses. Inhibiting integration with raltegravir shunted the generation of durable latency from integrated to unintegrated genomes. Latent uDNA was activated to de novo virus production by latency reversing agents that also activated latent integrated proviruses, including PKC activators, histone deacetylase inhibitors and P-TEFb agonists. However, uDNA responses displayed a wider dynamic range, indicating differential regulation of expression relative to integrated proviruses. Similar to what has recently been demonstrated for latent integrated proviruses, one or two applications of latency reversing agents failed to activate all latent unintegrated genomes. Unlike integrated proviruses, uDNA gene expression did not down modulate expression of HLA Class I on resting CD4 T cells. uDNA did, however, efficiently prime infected cells for killing by HIV-1-specific cytotoxic T cells.
Conclusions: These studies demonstrate that contributions by unintegrated genomes to HIV-1 gene expression, virus production, latency and immune responses are inherent properties of the direct infection of resting CD4 T cells. Experimental models of HIV-1 latency employing directly infected resting CD4 T cells should calibrate the contribution of unintegrated HIV-1.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

