Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer
- PMID: 26729235
- PMCID: PMC4700448
- DOI: 10.1038/srep18517
Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer
Abstract
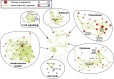
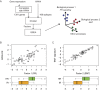
Invasive lobular carcinoma (ILC) is the second most frequently occurring histological breast cancer subtype after invasive ductal carcinoma (IDC), accounting for around 10% of all breast cancers. The molecular processes that drive the development of ILC are still largely unknown. We have performed a comprehensive genomic, transcriptomic and proteomic analysis of a large ILC patient cohort and present here an integrated molecular portrait of ILC. Mutations in CDH1 and in the PI3K pathway are the most frequent molecular alterations in ILC. We identified two main subtypes of ILCs: (i) an immune related subtype with mRNA up-regulation of PD-L1, PD-1 and CTLA-4 and greater sensitivity to DNA-damaging agents in representative cell line models; (ii) a hormone related subtype, associated with Epithelial to Mesenchymal Transition (EMT), and gain of chromosomes 1q and 8q and loss of chromosome 11q. Using the somatic mutation rate and eIF4B protein level, we identified three groups with different clinical outcomes, including a group with extremely good prognosis. We provide a comprehensive overview of the molecular alterations driving ILC and have explored links with therapy response. This molecular characterization may help to tailor treatment of ILC through the application of specific targeted, chemo- and/or immune-therapies.
Conflict of interest statement
W.M.G. is a co-founder and Chief Scientific Officer of OncoMark Limited. J.K.P., J.H., M.S. and I.M.S. received salary from Agendia as employee (no stocks or other funding). R.B. is employee and shareholder of Agendia. M.M., T.B., L.F.A.W. and R.B. are inventors on a patent about the ILC subtypes presented in this study.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

