The Art of Destruction: Optimizing Collision Energies in Quadrupole-Time of Flight (Q-TOF) Instruments for Glycopeptide-Based Glycoproteomics
- PMID: 26729457
- PMCID: PMC4756043
- DOI: 10.1007/s13361-015-1308-6
The Art of Destruction: Optimizing Collision Energies in Quadrupole-Time of Flight (Q-TOF) Instruments for Glycopeptide-Based Glycoproteomics
Abstract
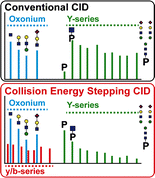
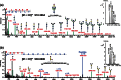
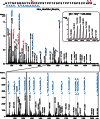
In-depth site-specific investigations of protein glycosylation are the basis for understanding the biological function of glycoproteins. Mass spectrometry-based N- and O-glycopeptide analyses enable determination of the glycosylation site, site occupancy, as well as glycan varieties present on a particular site. However, the depth of information is highly dependent on the applied analytical tools, including glycopeptide fragmentation regimes and automated data analysis. Here, we used a small set of synthetic disialylated, biantennary N-glycopeptides to systematically tune Q-TOF instrument parameters towards optimal energy stepping collision induced dissociation (CID) of glycopeptides. A linear dependency of m/z-ratio and optimal fragmentation energy was found, showing that with increasing m/z-ratio, more energy is required for glycopeptide fragmentation. Based on these optimized fragmentation parameters, a method combining lower- and higher-energy CID was developed, allowing the online acquisition of glycan and peptide-specific fragments within a single tandem MS experiment. We validated this method analyzing a set of human immunoglobulins (IgA1+2, sIgA, IgG1+2, IgE, IgD, IgM) as well as bovine fetuin. These optimized fragmentation parameters also enabled software-assisted glycopeptide assignment of both N- and O-glycopeptides including information about the most abundant glycan compositions, peptide sequence and putative structures. Twenty-six out of 30 N-glycopeptides and four out of five O-glycopeptides carrying >110 different glycoforms could be identified by this optimized LC-ESI tandem MS method with minimal user input. The Q-TOF based glycopeptide analysis platform presented here opens the way to a range of different applications in glycoproteomics research as well as biopharmaceutical development and quality control.
Keywords: Collision energy stepping CID; Glycopeptide; Glycoproteomics; Immunoglobulin; N-glycan; O-glycan; Q-TOF; Synthetic glycopeptides.
Figures


Similar articles
-
Site-Specific N- and O-Glycopeptide Analysis Using an Integrated C18-PGC-LC-ESI-QTOF-MS/MS Approach.Methods Mol Biol. 2017;1503:109-119. doi: 10.1007/978-1-4939-6493-2_9. Methods Mol Biol. 2017. PMID: 27743362
-
Glycoproteomics based on tandem mass spectrometry of glycopeptides.J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Apr 15;849(1-2):115-28. doi: 10.1016/j.jchromb.2006.09.041. Epub 2006 Oct 17. J Chromatogr B Analyt Technol Biomed Life Sci. 2007. PMID: 17049937 Review.
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
-
Determination of glycopeptide structures by multistage mass spectrometry with low-energy collision-induced dissociation: comparison of electrospray ionization quadrupole ion trap and matrix-assisted laser desorption/ionization quadrupole ion trap reflectron time-of-flight approaches.Rapid Commun Mass Spectrom. 2004;18(14):1575-82. doi: 10.1002/rcm.1521. Rapid Commun Mass Spectrom. 2004. PMID: 15282782
-
Parallel Determination of Polypeptide and Oligosaccharide Connectivities by Energy-Resolved Collison-Induced Dissociation of Protonated O-Glycopeptides Derived from Nonspecific Proteolysis.J Am Soc Mass Spectrom. 2020 Mar 4;31(3):624-632. doi: 10.1021/jasms.9b00065. Epub 2020 Feb 21. J Am Soc Mass Spectrom. 2020. PMID: 32126781 Free PMC article.
Cited by
-
Oxonium Ion Guided Analysis of Quantitative Proteomics Data Reveals Site-Specific O-Glycosylation of Anterior Gradient Protein 2 (AGR2).Int J Mol Sci. 2021 May 20;22(10):5369. doi: 10.3390/ijms22105369. Int J Mol Sci. 2021. PMID: 34065225 Free PMC article.
-
Can We Boost N-Glycopeptide Identification Confidence? Smart Collision Energy Choice Taking into Account Structure and Search Engine.J Am Soc Mass Spectrom. 2024 Feb 7;35(2):333-343. doi: 10.1021/jasms.3c00375. Epub 2024 Jan 29. J Am Soc Mass Spectrom. 2024. PMID: 38286027 Free PMC article.
-
Glycoproteomics Technologies in Glycobiotechnology.Adv Biochem Eng Biotechnol. 2021;175:413-434. doi: 10.1007/10_2020_144. Adv Biochem Eng Biotechnol. 2021. PMID: 33205259
-
Why Glycosylation Matters in Building a Better Flu Vaccine.Mol Cell Proteomics. 2019 Dec;18(12):2348-2358. doi: 10.1074/mcp.R119.001491. Epub 2019 Oct 11. Mol Cell Proteomics. 2019. PMID: 31604803 Free PMC article. Review.
-
Large-Scale Identification and Fragmentation Pathways Analysis of N-Glycans from Mouse Brain.J Am Soc Mass Spectrom. 2019 Jul;30(7):1254-1261. doi: 10.1007/s13361-019-02181-y. Epub 2019 May 16. J Am Soc Mass Spectrom. 2019. PMID: 31098956
References
-
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR. Essentials of Glycobiology. 2. New York: Cold Spring Harbor Laboratory Press; 2009. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

