Evolution of Pseudomonas aeruginosa Antimicrobial Resistance and Fitness under Low and High Mutation Rates
- PMID: 26729493
- PMCID: PMC4775977
- DOI: 10.1128/AAC.02676-15
Evolution of Pseudomonas aeruginosa Antimicrobial Resistance and Fitness under Low and High Mutation Rates
Abstract
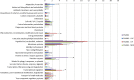
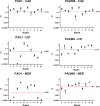
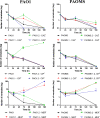
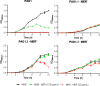
Pseudomonas aeruginosa, a major cause of nosocomial and chronic infections, is considered a paradigm of antimicrobial resistance development. However, the evolutionary trajectories of antimicrobial resistance and the impact of mutator phenotypes remain mostly unexplored. Therefore, whole-genome sequencing (WGS) was performed in lineages of wild-type and mutator (ΔmutS) strains exposed to increasing concentrations of relevant antipseudomonal agents. WGS provided a privileged perspective of the dramatic effect of mutator phenotypes on the accumulation of random mutations, most of which were transitions, as expected. Moreover, a frameshift mutagenic signature, consistent with error-prone DNA polymerase activity as a consequence of SOS system induction, was also seen. This effect was evidenced for all antibiotics tested, but it was higher for fluoroquinolones than for cephalosporins or carbapenems. Analysis of genotype versus phenotype confirmed expected resistance evolution trajectories but also revealed new pathways. Classical mechanisms included multiple mutations leading to AmpC overexpression (ceftazidime), quinolone resistance-determining region (QRDR) mutations (ciprofloxacin), oprD inactivation (meropenem), and efflux pump overexpression (ciprofloxacin and meropenem). Groundbreaking findings included gain-of-function mutations leading to the structural modification of AmpC (ceftazidime), novel DNA gyrase (GyrA) modification (ciprofloxacin), and the alteration of the β-lactam binding site of penicillin-binding protein 3 (PBP3) (meropenem). A further striking finding was seen in the evolution of meropenem resistance, selecting for specific extremely large (>250 kb) genomic deletions providing a growth advantage in the presence of the antibiotic. Finally, fitness and virulence varied within and across evolved antibiotic-resistant populations, but mutator lineages showed a lower biological cost for some antibiotics.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Vincent JL. 2003. Nosocomial infections in adult intensive-care units. Lancet 61:2068–2077. - PubMed
-
- Oliver A, Mena A, Macià MD. 2008. Evolution of Pseudomonas aeruginosa pathogenicity: from acute to chronic infections, p 433–444. In Baquero F, Nombela C, Cassell GH, Gutíerrez JA (ed), Evolutionary biology of bacterial and fungal pathogens. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

