Proteome-wide Identification of Novel Ceramide-binding Proteins by Yeast Surface cDNA Display and Deep Sequencing
- PMID: 26729710
- PMCID: PMC4824852
- DOI: 10.1074/mcp.M115.055954
Proteome-wide Identification of Novel Ceramide-binding Proteins by Yeast Surface cDNA Display and Deep Sequencing
Abstract
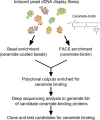
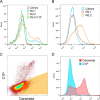
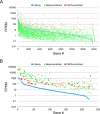
Although the bioactive sphingolipid ceramide is an important cell signaling molecule, relatively few direct ceramide-interacting proteins are known. We used an approach combining yeast surface cDNA display and deep sequencing technology to identify novel proteins binding directly to ceramide. We identified 234 candidate ceramide-binding protein fragments and validated binding for 20. Most (17) bound selectively to ceramide, although a few (3) bound to other lipids as well. Several novel ceramide-binding domains were discovered, including the EF-hand calcium-binding motif, the heat shock chaperonin-binding motif STI1, the SCP2 sterol-binding domain, and the tetratricopeptide repeat region motif. Interestingly, four of the verified ceramide-binding proteins (HPCA, HPCAL1, NCS1, and VSNL1) and an additional three candidate ceramide-binding proteins (NCALD, HPCAL4, and KCNIP3) belong to the neuronal calcium sensor family of EF hand-containing proteins. We used mutagenesis to map the ceramide-binding site in HPCA and to create a mutant HPCA that does not bind to ceramide. We demonstrated selective binding to ceramide by mammalian cell-produced wild type but not mutant HPCA. Intriguingly, we also identified a fragment from prostaglandin D2synthase that binds preferentially to ceramide 1-phosphate. The wide variety of proteins and domains capable of binding to ceramide suggests that many of the signaling functions of ceramide may be regulated by direct binding to these proteins. Based on the deep sequencing data, we estimate that our yeast surface cDNA display library covers ∼60% of the human proteome and our selection/deep sequencing protocol can identify target-interacting protein fragments that are present at extremely low frequency in the starting library. Thus, the yeast surface cDNA display/deep sequencing approach is a rapid, comprehensive, and flexible method for the analysis of protein-ligand interactions, particularly for the study of non-protein ligands.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Galadari S., Rahman A., Pallichankandy S., and Thayyullathil F. (2015) Tumor suppressive functions of ceramide: evidence and mechanisms. Apoptosis 20, 689–711 - PubMed
-
- Morad S. A., and Cabot M. C. (2013) Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 13, 51–65 - PubMed
-
- Saddoughi S. A., and Ogretmen B. (2013) Diverse functions of ceramide in cancer cell death and proliferation. Adv. Cancer Res. 117, 37–58 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

