Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence
- PMID: 26729876
- PMCID: PMC4725510
- DOI: 10.1073/pnas.1523321113
Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence
Abstract
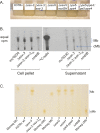
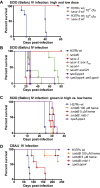
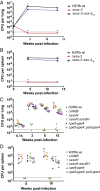
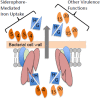
Mycobacterium tuberculosis (Mtb) encodes five type VII secretion systems (T7SS), designated ESX-1-ESX-5, that are critical for growth and pathogenesis. The best characterized is ESX-1, which profoundly impacts host cell interactions. In contrast, the ESX-3 T7SS is implicated in metal homeostasis, but efforts to define its function have been limited by an inability to recover deletion mutants. We overcame this impediment using medium supplemented with various iron complexes to recover mutants with deletions encompassing select genes within esx-3 or the entire operon. The esx-3 mutants were defective in uptake of siderophore-bound iron and dramatically accumulated cell-associated mycobactin siderophores. Proteomic analyses of culture filtrate revealed that secretion of EsxG and EsxH was codependent and that EsxG-EsxH also facilitated secretion of several members of the proline-glutamic acid (PE) and proline-proline-glutamic acid (PPE) protein families (named for conserved PE and PPE N-terminal motifs). Substrates that depended on EsxG-EsxH for secretion included PE5, encoded within the esx-3 locus, and the evolutionarily related PE15-PPE20 encoded outside the esx-3 locus. In vivo characterization of the mutants unexpectedly showed that the ESX-3 secretion system plays both iron-dependent and -independent roles in Mtb pathogenesis. PE5-PPE4 was found to be critical for the siderophore-mediated iron-acquisition functions of ESX-3. The importance of this iron-acquisition function was dependent upon host genotype, suggesting a role for ESX-3 secretion in counteracting host defense mechanisms that restrict iron availability. Further, we demonstrate that the ESX-3 T7SS secretes certain effectors that are important for iron uptake while additional secreted effectors modulate virulence in an iron-independent fashion.
Keywords: ESX-3; Mycobacterium tuberculosis; mycobactin; siderophore; type VII secretion system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Stoop EJ, Bitter W, van der Sar AM. Tubercle bacilli rely on a type VII army for pathogenicity. Trends Microbiol. 2012;20(10):477–484. - PubMed
-
- van der Woude AD, Luirink J, Bitter W. Getting across the cell envelope: Mycobacterial protein secretion. Curr Top Microbiol Immunol. 2013;374:109–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

