Epidemiologic studies of the human microbiome and cancer
- PMID: 26730578
- PMCID: PMC4742587
- DOI: 10.1038/bjc.2015.465
Epidemiologic studies of the human microbiome and cancer
Abstract
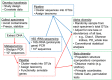
The human microbiome, which includes the collective genome of all bacteria, archaea, fungi, protists, and viruses found in and on the human body, is altered in many diseases and may substantially affect cancer risk. Previously detected associations of individual bacteria (e.g., Helicobacter pylori), periodontal disease, and inflammation with specific cancers have motivated studies considering the association between the human microbiome and cancer risk. This short review summarises microbiome research, focusing on published epidemiological associations with gastric, oesophageal, hepatobiliary, pancreatic, lung, colorectal, and other cancers. Large, prospective studies of the microbiome that employ multidisciplinary laboratory and analysis methods, as well as rigorous validation of case status, are likely to yield translational opportunities to reduce cancer morbidity and mortality by improving prevention, screening, and treatment.
Figures
References
-
- Abnet CC, Qiao YL, Mark SD, Dong ZW, Taylor PR, Dawsey SM (2001) Prospective study of tooth loss and incident esophageal and gastric cancers in China. Cancer Causes Control 12(9): 847–854. - PubMed
-
- Aigelsreiter A, Gerlza T, Deutsch AJ, Leitner E, Beham-Schmid C, Beham A, Popper H, Borel N, Pospischil A, Raderer M, Kessler HH, Neumeister P (2011) Chlamydia psittaci infection in nongastrointestinal extranodal MALT lymphomas and their precursor lesions. Am J Clin Pathol 135(1): 70–75. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

