TALENs Facilitate Single-step Seamless SDF Correction of F508del CFTR in Airway Epithelial Submucosal Gland Cell-derived CF-iPSCs
- PMID: 26730810
- PMCID: PMC5012545
- DOI: 10.1038/mtna.2015.43
TALENs Facilitate Single-step Seamless SDF Correction of F508del CFTR in Airway Epithelial Submucosal Gland Cell-derived CF-iPSCs
Abstract
Cystic fibrosis (CF) is a recessive inherited disease associated with multiorgan damage that compromises epithelial and inflammatory cell function. Induced pluripotent stem cells (iPSCs) have significantly advanced the potential of developing a personalized cell-based therapy for diseases like CF by generating patient-specific stem cells that can be differentiated into cells that repair tissues damaged by disease pathology. The F508del mutation in airway epithelial cell-derived CF-iPSCs was corrected with small/short DNA fragments (SDFs) and sequence-specific TALENs. An allele-specific PCR, cyclic enrichment strategy gave ~100-fold enrichment of the corrected CF-iPSCs after six enrichment cycles that facilitated isolation of corrected clones. The seamless SDF-based gene modification strategy used to correct the CF-iPSCs resulted in pluripotent cells that, when differentiated into endoderm/airway-like epithelial cells showed wild-type (wt) airway epithelial cell cAMP-dependent Cl ion transport or showed the appropriate cell-type characteristics when differentiated along mesoderm/hematopoietic inflammatory cell lineage pathways.
Figures
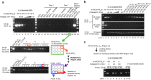
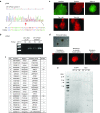
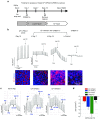
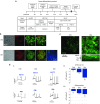
Similar articles
-
Augmentation of Cystic Fibrosis Transmembrane Conductance Regulator Function in Human Bronchial Epithelial Cells via SLC6A14-Dependent Amino Acid Uptake. Implications for Treatment of Cystic Fibrosis.Am J Respir Cell Mol Biol. 2019 Dec;61(6):755-764. doi: 10.1165/rcmb.2019-0094OC. Am J Respir Cell Mol Biol. 2019. PMID: 31189070
-
High-Efficiency, Selection-free Gene Repair in Airway Stem Cells from Cystic Fibrosis Patients Rescues CFTR Function in Differentiated Epithelia.Cell Stem Cell. 2020 Feb 6;26(2):161-171.e4. doi: 10.1016/j.stem.2019.11.002. Epub 2019 Dec 12. Cell Stem Cell. 2020. PMID: 31839569 Free PMC article.
-
Correction of Airway Stem Cells: Genome Editing Approaches for the Treatment of Cystic Fibrosis.Hum Gene Ther. 2020 Sep;31(17-18):956-972. doi: 10.1089/hum.2020.160. Epub 2020 Sep 8. Hum Gene Ther. 2020. PMID: 32741223 Free PMC article. Review.
-
Prime editing-mediated correction of the CFTR W1282X mutation in iPSCs and derived airway epithelial cells.PLoS One. 2023 Nov 29;18(11):e0295009. doi: 10.1371/journal.pone.0295009. eCollection 2023. PLoS One. 2023. PMID: 38019847 Free PMC article.
-
Induced pluripotent stem cells for treating cystic fibrosis: State of the science.Pediatr Pulmonol. 2018 Nov;53(S3):S12-S29. doi: 10.1002/ppul.24118. Epub 2018 Jul 30. Pediatr Pulmonol. 2018. PMID: 30062693 Review.
Cited by
-
Seamless Gene Correction in the Human Cystic Fibrosis Transmembrane Conductance Regulator Locus by Vector Replacement and Vector Insertion Events.Front Genome Ed. 2022 Apr 6;4:843885. doi: 10.3389/fgeed.2022.843885. eCollection 2022. Front Genome Ed. 2022. PMID: 35465025 Free PMC article.
-
iPSC-Derived Intestinal Organoids from Cystic Fibrosis Patients Acquire CFTR Activity upon TALEN-Mediated Repair of the p.F508del Mutation.Mol Ther Methods Clin Dev. 2020 Apr 18;17:858-870. doi: 10.1016/j.omtm.2020.04.005. eCollection 2020 Jun 12. Mol Ther Methods Clin Dev. 2020. PMID: 32373648 Free PMC article.
-
Induced Pluripotent Stem Cells Meet Genome Editing.Cell Stem Cell. 2016 May 5;18(5):573-86. doi: 10.1016/j.stem.2016.04.013. Cell Stem Cell. 2016. PMID: 27152442 Free PMC article. Review.
-
Airway regeneration using iPS cell-derived airway epithelial cells with Cl- channel function.Channels (Austin). 2019 Dec;13(1):227-234. doi: 10.1080/19336950.2019.1628550. Channels (Austin). 2019. PMID: 31198082 Free PMC article. Review.
-
Combining Induced Pluripotent Stem Cells and Genome Editing Technologies for Clinical Applications.Cell Transplant. 2018 Mar;27(3):379-392. doi: 10.1177/0963689718754560. Epub 2018 May 28. Cell Transplant. 2018. PMID: 29806481 Free PMC article.
References
-
- Riordan, JR, Rommens, JM, Kerem, B, Alon, N, Rozmahel, R, Grzelczak, Z et al. (1989). Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245: 1066–1073. - PubMed
-
- Consortium (2009). Cystic Fibrosis Mutation Database<http://www.genet.sickkids.on.ca/cftr/>.
-
- Andersson, C, Zaman, MM, Jones, AB and Freedman, SD (2008). Alterations in immune response and PPAR/LXR regulation in cystic fibrosis macrophages. J Cyst Fibros 7: 68–78. - PubMed
-
- Hartl, D, Gaggar, A, Bruscia, E, Hector, A, Marcos, V, Jung, A et al. (2012). Innate immunity in cystic fibrosis lung disease. J Cyst Fibros 11: 363–382. - PubMed
-
- Takahashi, K, Tanabe, K, Ohnuki, M, Narita, M, Ichisaka, T, Tomoda, K et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

