Decoding Biomass-Sensing Regulons of Clostridium thermocellum Alternative Sigma-I Factors in a Heterologous Bacillus subtilis Host System
- PMID: 26731480
- PMCID: PMC4711584
- DOI: 10.1371/journal.pone.0146316
Decoding Biomass-Sensing Regulons of Clostridium thermocellum Alternative Sigma-I Factors in a Heterologous Bacillus subtilis Host System
Abstract
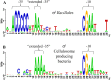
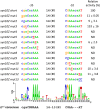
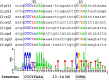
The Gram-positive, anaerobic, cellulolytic, thermophile Clostridium (Ruminiclostridium) thermocellum secretes a multi-enzyme system called the cellulosome to solubilize plant cell wall polysaccharides. During the saccharolytic process, the enzymatic composition of the cellulosome is modulated according to the type of polysaccharide(s) present in the environment. C. thermocellum has a set of eight alternative RNA polymerase sigma (σ) factors that are activated in response to extracellular polysaccharides and share sequence similarity to the Bacillus subtilis σI factor. The aim of the present work was to demonstrate whether individual C. thermocellum σI-like factors regulate specific cellulosomal genes, focusing on C. thermocellum σI6 and σI3 factors. To search for putative σI6- and σI3-dependent promoters, bioinformatic analysis of the upstream regions of the cellulosomal genes was performed. Because of the limited genetic tools available for C. thermocellum, the functionality of the predicted σI6- and σI3-dependent promoters was studied in B. subtilis as a heterologous host. This system enabled observation of the activation of 10 predicted σI6-dependent promoters associated with the C. thermocellum genes: sigI6 (itself, Clo1313_2778), xyn11B (Clo1313_0522), xyn10D (Clo1313_0177), xyn10Z (Clo1313_2635), xyn10Y (Clo1313_1305), cel9V (Clo1313_0349), cseP (Clo1313_2188), sigI1 (Clo1313_2174), cipA (Clo1313_0627), and rsgI5 (Clo1313_0985). Additionally, we observed the activation of 4 predicted σI3-dependent promoters associated with the C. thermocellum genes: sigI3 (itself, Clo1313_1911), pl11 (Clo1313_1983), ce12 (Clo1313_0693) and cipA. Our results suggest possible regulons of σI6 and σI3 in C. thermocellum, as well as the σI6 and σI3 promoter consensus sequences. The proposed -35 and -10 promoter consensus elements of σI6 are CNNAAA and CGAA, respectively. Additionally, a less conserved CGA sequence next to the C in the -35 element and a highly conserved AT sequence three bases downstream of the -10 element were also identified as important nucleotides for promoter recognition. Regarding σI3, the proposed -35 and -10 promoter consensus elements are CCCYYAAA and CGWA, respectively. The present study provides new clues for understanding these recently discovered alternative σI factors.
Conflict of interest statement
Figures

References
-
- Viljoen JA, Fred EB, Peterson WH. The fermentation of cellulose by thermophilic bacteria. J Agric Sci. 1926; 16: 1–17. 10.1017/S0021859600088249 - DOI
-
- Lamed R, Setter E, Kenig R, Bayer EA. The cellulosome—a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol Bioeng Symp. 1983; 13: 163–181.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

