Transient Accumulation of NO2- and N2O during Denitrification Explained by Assuming Cell Diversification by Stochastic Transcription of Denitrification Genes
- PMID: 26731685
- PMCID: PMC4701171
- DOI: 10.1371/journal.pcbi.1004621
Transient Accumulation of NO2- and N2O during Denitrification Explained by Assuming Cell Diversification by Stochastic Transcription of Denitrification Genes
Abstract
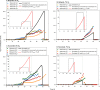
Denitrifying bacteria accumulate [Formula: see text], NO, and N2O, the amounts depending on transcriptional regulation of core denitrification genes in response to O2-limiting conditions. The genes include nar, nir, nor and nosZ, encoding [Formula: see text]-, [Formula: see text]-, NO- and N2O reductase, respectively. We previously constructed a dynamic model to simulate growth and respiration in batch cultures of Paracoccus denitrificans. The observed denitrification kinetics were adequately simulated by assuming a stochastic initiation of nir-transcription in each cell with an extremely low probability (0.5% h-1), leading to product- and substrate-induced transcription of nir and nor, respectively, via NO. Thus, the model predicted cell diversification: after O2 depletion, only a small fraction was able to grow by reducing [Formula: see text]. Here we have extended the model to simulate batch cultivation with [Formula: see text], i.e., [Formula: see text], NO, N2O, and N2 kinetics, measured in a novel experiment including frequent measurements of [Formula: see text]. Pa. denitrificans reduced practically all [Formula: see text] to [Formula: see text] before initiating gas production. The [Formula: see text] production is adequately simulated by assuming stochastic nar-transcription, as that for nirS, but with a higher probability (0.035 h-1) and initiating at a higher O2 concentration. Our model assumes that all cells express nosZ, thus predicting that a majority of cells have only N2O-reductase (A), while a minority (B) has [Formula: see text]-, NO- and N2O-reductase. Population B has a higher cell-specific respiration rate than A because the latter can only use N2O produced by B. Thus, the ratio [Formula: see text] is low immediately after O2 depletion, but increases throughout the anoxic phase because B grows faster than A. As a result, the model predicts initially low but gradually increasing N2O concentration throughout the anoxic phase, as observed. The modelled cell diversification neatly explains the observed denitrification kinetics and transient intermediate accumulations. The result has major implications for understanding the relationship between genotype and phenotype in denitrification research.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Bates Bryson, Kundzewicz Zbigniew W., Wu Shaohong, Arnell Nigel, Burkett Virginia, et al. (2008) Technical paper on climate change and water Budapest: Intergovernmental Panel on Climate Change;
-
- Lassey K, Harvey M (2007) Nitrous oxide: the serious side of laughing gas. Water & Atmos 15: 10–11.
-
- Signor D, Cerri CEP (2013) Nitrous oxide emissions in agricultural soils: a review. Pesq Agropec Trop 43: 322–338.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

