Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics
- PMID: 26732734
- PMCID: PMC4729823
- DOI: 10.1038/ncomms10261
Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics
Abstract
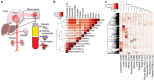
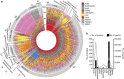
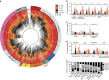
The plasma proteome is highly dynamic and variable, composed of proteins derived from surrounding tissues and cells. To investigate the complex processes that control the composition of the plasma proteome, we developed a mass spectrometry-based proteomics strategy to infer the origin of proteins detected in murine plasma. The strategy relies on the construction of a comprehensive protein tissue atlas from cells and highly vascularized organs using shotgun mass spectrometry. The protein tissue atlas was transformed to a spectral library for highly reproducible quantification of tissue-specific proteins directly in plasma using SWATH-like data-independent mass spectrometry analysis. We show that the method can determine drastic changes of tissue-specific protein profiles in blood plasma from mouse animal models with sepsis. The strategy can be extended to several other species advancing our understanding of the complex processes that contribute to the plasma proteome dynamics.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

