Identification of Proteins Bound to Dengue Viral RNA In Vivo Reveals New Host Proteins Important for Virus Replication
- PMID: 26733069
- PMCID: PMC4725007
- DOI: 10.1128/mBio.01865-15
Identification of Proteins Bound to Dengue Viral RNA In Vivo Reveals New Host Proteins Important for Virus Replication
Abstract
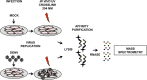
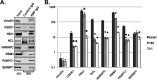
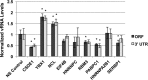
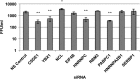
Dengue virus is the most prevalent cause of arthropod-borne infection worldwide. Due to the limited coding capacity of the viral genome and the complexity of the viral life cycle, host cell proteins play essential roles throughout the course of viral infection. Host RNA-binding proteins mediate various aspects of virus replication through their physical interactions with viral RNA. Here we describe a technique designed to identify such interactions in the context of infected cells using UV cross-linking followed by antisense-mediated affinity purification and mass spectrometry. Using this approach, we identified interactions, several of them novel, between host proteins and dengue viral RNA in infected Huh7 cells. Most of these interactions were subsequently validated using RNA immunoprecipitation. Using small interfering RNA (siRNA)-mediated gene silencing, we showed that more than half of these host proteins are likely involved in regulating virus replication, demonstrating the utility of this method in identifying biologically relevant interactions that may not be identified using traditional in vitro approaches.
Importance: Dengue virus is the most prevalent cause of arthropod-borne infection worldwide. Viral RNA molecules physically interact with cellular RNA-binding proteins (RBPs) throughout the course of infection; the identification of such interactions will lead to the elucidation of the molecular mechanisms of virus replication. Until now, the identification of host proteins bound to dengue viral RNA has been accomplished using in vitro strategies. Here, we used a method for the specific purification of dengue viral ribonucleoprotein (RNP) complexes from infected cells and subsequently identified the associated proteins by mass spectrometry. We then validated a functional role for the majority of these proteins in mediating efficient virus replication. This approach has broad relevance to virology and RNA biology, as it could theoretically be used to purify any viral RNP complex of interest.
Copyright © 2016 Phillips et al.
Figures

References
-
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi:10.1038/nature12060. - DOI - PMC - PubMed
-
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin T, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Shi P, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi:10.1038/nature09489. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

