Increased IGFBP-1 phosphorylation in response to leucine deprivation is mediated by CK2 and PKC
- PMID: 26733150
- PMCID: PMC4811673
- DOI: 10.1016/j.mce.2015.12.006
Increased IGFBP-1 phosphorylation in response to leucine deprivation is mediated by CK2 and PKC
Abstract
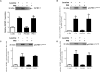
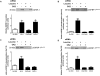
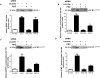
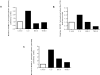
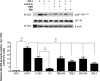
Insulin-like growth factor binding protein-1 (IGFBP-1), secreted by fetal liver, is a key regulator of IGF-I bioavailability and fetal growth. IGFBP-1 phosphorylation decreases IGF-I bioavailability and diminishes its growth-promoting effects. Growth-restricted fetuses have decreased levels of circulating essential amino acids. We recently showed that IGFBP-1 hyperphosphorylation (pSer101/119/169) in response to leucine deprivation is regulated via activation of the amino acid response (AAR) in HepG2 cells. Here we investigated nutrient-sensitive protein kinases CK2/PKC/PKA in mediating IGFBP-1 phosphorylation in leucine deprivation. We demonstrated that leucine deprivation stimulated CK2 activity (enzymatic assay) and induced IGFBP-1 phosphorylation (immunoblotting/MRM-MS). Inhibition (pharmacological/siRNA) of CK2/PKC, but not PKA, prevented IGFBP-1 hyperphosphorylation in leucine deprivation. PKC inhibition also prevented leucine deprivation-stimulated CK2 activity. Functionally, leucine deprivation decreased IGF-I-induced-IGF-1R autophosphorylation when CK2/PKC were not inhibited. Our data strongly support that PKC promotes leucine deprivation-induced IGFBP-1 hyperphosphorylation via CK2 activation, mechanistically linking decreased amino acid availability and reduced fetal growth.
Keywords: Amino acid restriction; Fetal growth; HepG2 cells; Insulin-like growth Factor-1 receptor; Insulin-like growth factor binding protein; Mass spectrometry; Phosphorylation sites; Protein kinases.
Copyright © 2015 Elsevier Ireland Ltd. All rights reserved.
Figures



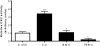
Similar articles
-
IGFBP-1 hyperphosphorylation in response to leucine deprivation is mediated by the AAR pathway.Mol Cell Endocrinol. 2015 Sep 5;412:182-95. doi: 10.1016/j.mce.2015.04.031. Epub 2015 May 5. Mol Cell Endocrinol. 2015. PMID: 25957086 Free PMC article.
-
Liver mTOR controls IGF-I bioavailability by regulation of protein kinase CK2 and IGFBP-1 phosphorylation in fetal growth restriction.Endocrinology. 2014 Apr;155(4):1327-39. doi: 10.1210/en.2013-1759. Epub 2014 Jan 17. Endocrinology. 2014. PMID: 24437487 Free PMC article.
-
Inhibition of decidual IGF-1 signaling in response to hypoxia and leucine deprivation is mediated by mTOR and AAR pathways and increased IGFBP-1 phosphorylation.Mol Cell Endocrinol. 2020 Jul 15;512:110865. doi: 10.1016/j.mce.2020.110865. Epub 2020 Jun 5. Mol Cell Endocrinol. 2020. PMID: 32502935
-
Insulin-like growth factor binding protein-1: recent findings and new directions.Proc Soc Exp Biol Med. 1997 Dec;216(3):319-57. doi: 10.3181/00379727-216-44182. Proc Soc Exp Biol Med. 1997. PMID: 9402139 Review.
-
Phosphorylation of insulin-like growth factor binding proteins.Mol Cell Endocrinol. 1997 Apr 4;128(1-2):1-5. doi: 10.1016/s0303-7207(97)04032-x. Mol Cell Endocrinol. 1997. PMID: 9140069 Review.
Cited by
-
IGFBP-1 hyperphosphorylation in response to nutrient deprivation is mediated by activation of protein kinase Cα (PKCα).Mol Cell Endocrinol. 2021 Oct 1;536:111400. doi: 10.1016/j.mce.2021.111400. Epub 2021 Jul 24. Mol Cell Endocrinol. 2021. PMID: 34314739 Free PMC article.
-
Hyperphosphorylation of fetal liver IGFBP-1 precedes slowing of fetal growth in nutrient-restricted baboons and may be a mechanism underlying IUGR.Am J Physiol Endocrinol Metab. 2020 Sep 1;319(3):E614-E628. doi: 10.1152/ajpendo.00220.2020. Epub 2020 Aug 3. Am J Physiol Endocrinol Metab. 2020. PMID: 32744097 Free PMC article.
-
Identification of PKCα-dependent phosphoproteins in mouse retina.J Proteomics. 2019 Aug 30;206:103423. doi: 10.1016/j.jprot.2019.103423. Epub 2019 Jun 28. J Proteomics. 2019. PMID: 31255707 Free PMC article.
-
Increased Colocalization and Interaction Between Decidual Protein Kinase A and Insulin-like Growth Factor-Binding Protein-1 in Intrauterine Growth Restriction.J Histochem Cytochem. 2022 Jul;70(7):515-530. doi: 10.1369/00221554221112702. Epub 2022 Jul 8. J Histochem Cytochem. 2022. PMID: 35801847 Free PMC article.
-
Novel roles of mechanistic target of rapamycin signaling in regulating fetal growth†.Biol Reprod. 2019 Apr 1;100(4):872-884. doi: 10.1093/biolre/ioy249. Biol Reprod. 2019. PMID: 30476008 Free PMC article. Review.
References
-
- Abu Shehab M, Iosef C, Wildgruber R, Sardana G, Gupta MB. Phosphorylation of IGFBP-1 at discrete sites elicits variable effects on IGF-I receptor autophosphorylation. Endocrinology. 2013;154:1130–1143. - PubMed
-
- Abu Shehab M, Khosravi J, Han VK, Shilton BH, Gupta MB. Site-specific IGFBP-1 hyper-phosphorylation in fetal growth restriction: clinical and functional relevance. J. Proteome Res. 2010;9:1873–1881. - PubMed
-
- Ankrapp DP, Jones JI, Clemmons DR. Characterization of insulin-like growth factor binding protein-1 kinases from human hepatoma cells. J. Cell. Biochem. 1996;60:387–399. - PubMed
-
- Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

