Base and Nucleotide Excision Repair of Oxidatively Generated Guanine Lesions in DNA
- PMID: 26733197
- PMCID: PMC4777862
- DOI: 10.1074/jbc.M115.693218
Base and Nucleotide Excision Repair of Oxidatively Generated Guanine Lesions in DNA
Abstract
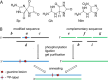
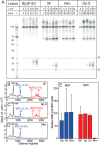
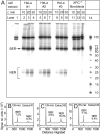
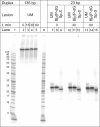
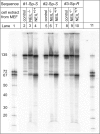
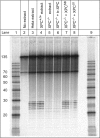
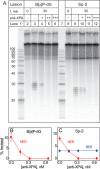
The well known biomarker of oxidative stress, 8-oxo-7,8-dihydroguanine, is more susceptible to further oxidation than the parent guanine base and can be oxidatively transformed to the genotoxic spiroiminodihydantoin (Sp) and 5-guanidinohydantoin (Gh) lesions. Incubation of 135-mer duplexes with single Sp or Gh lesions in human cell extracts yields a characteristic nucleotide excision repair (NER)-induced ladder of short dual incision oligonucleotide fragments in addition to base excision repair (BER) incision products. The ladders were not observed when NER was inhibited either by mouse monoclonal antibody (5F12) to human XPA or in XPC(-/-) fibroblast cell extracts. However, normal NER activity appeared when the XPC(-/-) cell extracts were complemented with XPC-RAD23B proteins. The Sp and Gh lesions are excellent substrates of both BER and NER. In contrast, 5-guanidino-4-nitroimidazole, a product of the oxidation of guanine in DNA by peroxynitrite, is an excellent substrate of BER only. In the case of mouse embryonic fibroblasts, BER of the Sp lesion is strongly reduced in NEIL1(-/-) relative to NEIL1(+/+) extracts. In summary, in human cell extracts, BER and NER activities co-exist and excise Gh and Sp DNA lesions, suggesting that the relative NER/BER product ratios may depend on competitive BER and NER protein binding to these lesions.
Keywords: DNA damage; DNA repair; base excision repair (BER); nucleotide excision repair; oxidative stress.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Cadet J., Douki T., and Ravanat J. L. (2008) Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. Acc. Chem. Res. 41, 1075–1083 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

