Cancer Metabolism: A Modeling Perspective
- PMID: 26733270
- PMCID: PMC4679931
- DOI: 10.3389/fphys.2015.00382
Cancer Metabolism: A Modeling Perspective
Abstract
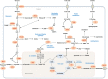
Tumor cells alter their metabolism to maintain unregulated cellular proliferation and survival, but this transformation leaves them reliant on constant supply of nutrients and energy. In addition to the widely studied dysregulated glucose metabolism to fuel tumor cell growth, accumulating evidences suggest that utilization of amino acids and lipids contributes significantly to cancer cell metabolism. Also recent progresses in our understanding of carcinogenesis have revealed that cancer is a complex disease and cannot be understood through simple investigation of genetic mutations of cancerous cells. Cancer cells present in complex tumor tissues communicate with the surrounding microenvironment and develop traits which promote their growth, survival, and metastasis. Decoding the full scope and targeting dysregulated metabolic pathways that support neoplastic transformations and their preservation requires both the advancement of experimental technologies for more comprehensive measurement of omics as well as the advancement of robust computational methods for accurate analysis of the generated data. Here, we review cancer-associated reprogramming of metabolism and highlight the capability of genome-scale metabolic modeling approaches in perceiving a system-level perspective of cancer metabolism and in detecting novel selective drug targets.
Keywords: cancer metabolism; genome scale metabolic reconstruction; metabolic modeling; metabolic networks and pathways; systems biology; systems medicine; tumor metabolism.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

