Helicobacter pylori ATCC 43629/NCTC 11639 Outer Membrane Vesicles (OMVs) from Biofilm and Planktonic Phase Associated with Extracellular DNA (eDNA)
- PMID: 26733944
- PMCID: PMC4679919
- DOI: 10.3389/fmicb.2015.01369
Helicobacter pylori ATCC 43629/NCTC 11639 Outer Membrane Vesicles (OMVs) from Biofilm and Planktonic Phase Associated with Extracellular DNA (eDNA)
Abstract
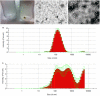
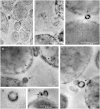
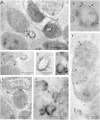
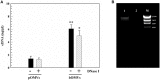
Helicobacter pylori persistence is associated with its capacity to develop biofilms as a response to changing environmental conditions and stress. Extracellular DNA (eDNA) is a component of H. pylori biofilm matrix but the lack of DNase I activity supports the hypothesis that eDNA might be protected by other extracellular polymeric substances (EPS) and/or Outer Membrane Vesicles (OMVs), which bleb from the bacteria surface during growth. The aim of the present study was to both identify the eDNA presence on OMVs segregated from H. pylori ATCC 43629/NCTC 11639 biofilm (bOMVs) and its planktonic phase (pOMVs) and to characterize the physical-chemical properties of the OMVs. The presence of eDNA in bOMVs and pOMVs was initially carried out using DNase I-gold complex labeling and Transmission Electron Microscope analysis (TEM). bOMVs and pOMVs were further isolated and physical-chemical characterization carried out using dynamic light scattering (DLS) analysis. eDNA associated with OMVs was detected and quantified using a PicoGreen spectrophotometer assay, while its extraction was performed with a DNA Kit. TEM images showed that eDNA was mainly associated with the OMV membrane surfaces; while PicoGreen staining showed a four-fold increase of dsDNA in bOMVs compared with pOMVs. The eDNA extracted from OMVs was visualized using gel electrophoresis. DLS analysis indicated that both planktonic and biofilm H. pylori phenotypes generated vesicles, with a broad distribution of sizes on the nanometer scale. The DLS aggregation assay suggested that eDNA may play a role in the aggregation of OMVs, in the biofilm phenotype. Moreover, the eDNA associated with vesicle membrane may impede DNase I activity on H. pylori biofilms. These results suggest that OMVs derived from the H. pylori biofilm phenotype may play a structural role by preventing eDNA degradation by nucleases, providing a bridging function between eDNA strands on OMV surfaces and promoting aggregation.
Keywords: Helicobacter pylori; biofilm; eDNA; nanoparticles; outer membrane vesicles (OMVs).
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

