Year-Long Metagenomic Study of River Microbiomes Across Land Use and Water Quality
- PMID: 26733955
- PMCID: PMC4681185
- DOI: 10.3389/fmicb.2015.01405
Year-Long Metagenomic Study of River Microbiomes Across Land Use and Water Quality
Abstract
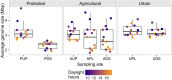
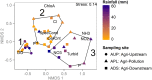
Select bacteria, such as Escherichia coli or coliforms, have been widely used as sentinels of low water quality; however, there are concerns regarding their predictive accuracy for the protection of human and environmental health. To develop improved monitoring systems, a greater understanding of bacterial community structure, function, and variability across time is required in the context of different pollution types, such as agricultural and urban contamination. Here, we present a year-long survey of free-living bacterial DNA collected from seven sites along rivers in three watersheds with varying land use in Southwestern Canada. This is the first study to examine the bacterial metagenome in flowing freshwater (lotic) environments over such a time span, providing an opportunity to describe bacterial community variability as a function of land use and environmental conditions. Characteristics of the metagenomic data, such as sequence composition and average genome size (AGS), vary with sampling site, environmental conditions, and water chemistry. For example, AGS was correlated with hours of daylight in the agricultural watershed and, across the agriculturally and urban-affected sites, k-mer composition clustering corresponded to nutrient concentrations. In addition to indicating a community shift, this change in AGS has implications in terms of the normalization strategies required, and considerations surrounding such strategies in general are discussed. When comparing abundances of gene functional groups between high- and low-quality water samples collected from an agricultural area, the latter had a higher abundance of nutrient metabolism and bacteriophage groups, possibly reflecting an increase in agricultural runoff. This work presents a valuable dataset representing a year of monthly sampling across watersheds and an analysis targeted at establishing a foundational understanding of how bacterial lotic communities vary across time and land use. The results provide important context for future studies, including further analyses of watershed ecosystem health, and the identification and development of biomarkers for improved water quality monitoring systems.
Keywords: bacteria; freshwater; land use; metagenomics; normalization; rivers; temporal variation.
Figures



References
-
- Aggarwal C. C., Hinneburg A., Keim D. A. (2001). On the surprising behavior of distance metrics in high dimensional spaces. Database Theory 1973 420–434.
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

