Comparative Genomics of Pathogenic and Nonpathogenic Strains of Xanthomonas arboricola Unveil Molecular and Evolutionary Events Linked to Pathoadaptation
- PMID: 26734033
- PMCID: PMC4686621
- DOI: 10.3389/fpls.2015.01126
Comparative Genomics of Pathogenic and Nonpathogenic Strains of Xanthomonas arboricola Unveil Molecular and Evolutionary Events Linked to Pathoadaptation
Abstract
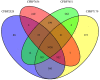
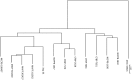
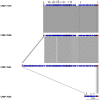
The bacterial species Xanthomonas arboricola contains plant pathogenic and nonpathogenic strains. It includes the pathogen X. arboricola pv. juglandis, causing the bacterial blight of Juglans regia. The emergence of a new bacterial disease of J. regia in France called vertical oozing canker (VOC) was previously described and the causal agent was identified as a distinct genetic lineage within the pathovar juglandis. Symptoms on walnut leaves and fruits are similar to those of a bacterial blight but VOC includes also cankers on trunk and branches. In this work, we used comparative genomics and physiological tests to detect differences between four X. arboricola strains isolated from walnut tree: strain CFBP 2528 causing walnut blight (WB), strain CFBP 7179 causing VOC and two nonpathogenic strains, CFBP 7634 and CFBP 7651, isolated from healthy walnut buds. Whole genome sequence comparisons revealed that pathogenic strains possess a larger and wider range of mobile genetic elements than nonpathogenic strains. One pathogenic strain, CFBP 7179, possessed a specific integrative and conjugative element (ICE) of 95 kb encoding genes involved in copper resistance, transport and regulation. The type three effector repertoire was larger in pathogenic strains than in nonpathogenic strains. Moreover, CFBP 7634 strain lacked the type three secretion system encoding genes. The flagellar system appeared incomplete and nonfunctional in the pathogenic strain CFBP 2528. Differential sets of chemoreceptor and different repertoires of genes coding adhesins were identified between pathogenic and nonpathogenic strains. Besides these differences, some strain-specific differences were also observed. Altogether, this study provides valuable insights to highlight the mechanisms involved in ecology, environment perception, plant adhesion and interaction, leading to the emergence of new strains in a dynamic environment.
Keywords: ICE; Juglans regia; bacterial blight; copper resistance; vertical oozing canker.
Figures




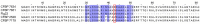

References
-
- Araki H., Tian D., Goss E. M., Jakob K., Halldorsdottir S. S., Kreitman M., et al. (2006). Presence/absence polymorphism for alternative pathogenicity islands in Pseudomonas viridiflava, a pathogen of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 5887–5892. 10.1073/pnas.0601431103 - DOI - PMC - PubMed
-
- Araújo E. R., Pereira R. C., Ferreira M. A. S. V., Fitopatologia D., Brasília U., De Hortaliças E., et al. (2012). Sensitivity of xanthomonads causing tomato bacterial spot to copper and streptomycin and in vivo infra-specific competitive ability in Xanthomonas perforans resistant and sensitive to copper. J. Plant Pathol. 94, 79–87. 10.4454/jpp.fa.2012.004 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

