RNA-Seq Analysis of Rice Roots Reveals the Involvement of Post-Transcriptional Regulation in Response to Cadmium Stress
- PMID: 26734039
- PMCID: PMC4685130
- DOI: 10.3389/fpls.2015.01136
RNA-Seq Analysis of Rice Roots Reveals the Involvement of Post-Transcriptional Regulation in Response to Cadmium Stress
Abstract
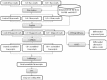
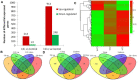
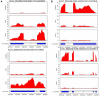
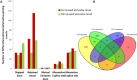
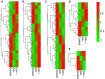
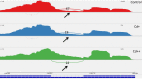
Widely-spread cadmium (Cd) pollution in the soil threatens both crop production and human health. How plants deal with the excess Cd are largely unknown. To evaluate the molecular mechanism by which plants respond to Cd stress, rice seedlings were treated with two concentrations of Cd and subjected to deep RNA sequencing. Comprehensive RNA-Seq analysis of rice roots under two gradients of Cd treatment revealed 1169 Cd toxicity-responsive genes. These genes were involved in the reactive oxygen species scavenging system, stress response, cell wall formation, ion transport, and signal transduction. Nine out of 93 predicted long non-coding RNAs (lncRNAs) were detected as Cd-responsive lncRNAs due to their high correlation with the Cd stress response. In addition, we analyzed alternative splicing (AS) events under different Cd concentrations. Four hundred and seventy-six differential alternatively spliced genes with 542 aberrant splicing events were identified. GO analysis indicated that these genes were highly enriched in oxidation reduction and cellular response to chemical stimulus. Real-time qRT-PCR validation analysis strengthened the reliability of our RNA-Seq results. The results suggest that post-transcriptional AS regulation may also be involved in plant responses to high Cd stress.
Keywords: RNA-seq; alternative splicing; cadmium; lncRNAs; rice root.
Figures





References
-
- Ali-Benali M. A., Badawi M., Houde Y., Houde M. (2013). Identification of oxidative stress-responsive C2H2 zinc fingers associated with Al tolerance in near-isogenic wheat lines. Plant Soil. 366, 199–212. 10.1007/s11104-012-1417-y - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

